8T18
 
 | |
8T15
 
 | |
8T17
 
 | |
8T6K
 
 | |
8T6Q
 
 | |
8SYG
 
 | |
3VRJ
 
 | | HLA-B*57:01-LTTKLTNTNI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
3VRI
 
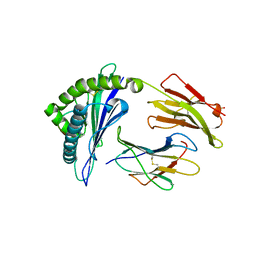 | | HLA-B*57:01-RVAQLENVYI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
4UV6
 
 | |
4H6S
 
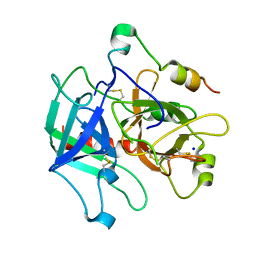 | | Crystal structure of thrombin mutant E14eA/D14lA/E18A/S195A | | Descriptor: | Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4HFP
 
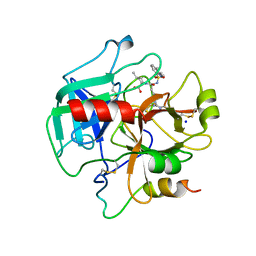 | | Structure of thrombin mutant S195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Lin, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4GI3
 
 | |
4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
3FPO
 
 | |
3FTH
 
 | |
3FTL
 
 | |
3FTK
 
 | |
3FOD
 
 | |
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
7YX9
 
 | |
