5TLW
 
 | |
5TEG
 
 | |
4Z3A
 
 | |
8EJM
 
 | |
8EHG
 
 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
5TI5
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841880 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
4Z7Z
 
 | | Structure of the enzyme-product complex resulting from TDG action on a GT mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z96
 
 | | Crystal structure of DNMT1 in complex with USP7 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.M, Song, J. | | Deposit date: | 2015-04-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of DNMT1 in complex with USP7
To Be Published
|
|
5TIJ
 
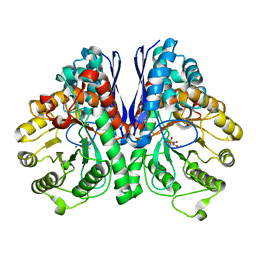 | | Structure of Human Enolase 2 with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonate (purified enantiomer) | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Eradication of ENO1-deleted Glioblastoma through Collateral Lethality
Biorxiv, 2019
|
|
5TI4
 
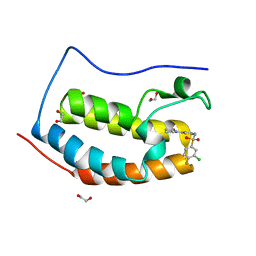 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841871 | | Descriptor: | 1,2-ETHANEDIOL, 3-chloro-4-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
4ZA0
 
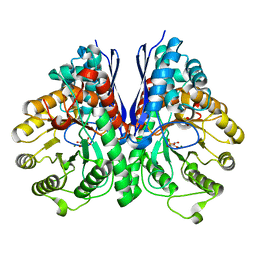 | | Structure of Human Enolase 2 in complex with Phosphonoacetohydroxamate | | Descriptor: | Gamma-enolase, MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
5VTB
 
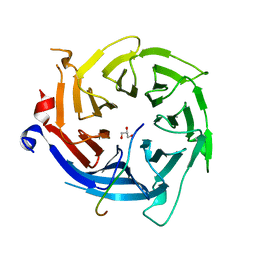 | | Crystal structure of RBBP4 bound to BCL11a peptide | | Descriptor: | B-cell lymphoma/leukemia 11A, GLYCEROL, Histone-binding protein RBBP4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes.
J. Biol. Chem., 293, 2018
|
|
4Z49
 
 | |
5UWR
 
 | |
5UPD
 
 | | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1) | | Descriptor: | Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Yu, W, Dong, A, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1)
To Be Published
|
|
4Z7B
 
 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
5VOM
 
 | | Benzopiperazine BET bromodomain inhibitor in complex with BD1 of Brd4 | | Descriptor: | 3-[(2S)-1-acetyl-4-(furan-2-carbonyl)-2-methyl-1,2,3,4-tetrahydroquinoxalin-6-yl]-N-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Toms, A.V, Herbertz, T. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design and Optimization of Benzopiperazines as Potent Inhibitors of BET Bromodomains.
ACS Med Chem Lett, 8, 2017
|
|
5T2W
 
 | |
5VY5
 
 | | Rabbit muscle aldolase using 200keV | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Herzik Jr, M.A, Wu, M, Lander, G.C. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Achieving better-than-3- angstrom resolution by single-particle cryo-EM at 200 keV.
Nat. Methods, 14, 2017
|
|
5VZS
 
 | | BRD4-BD1 in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, Bromodomain-containing protein 4 | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-29 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of CBP/P300
To be published
|
|
3I7O
 
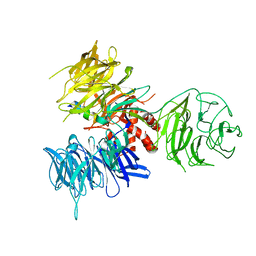 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of IQWD1 | | Descriptor: | DNA damage-binding protein 1, IQ motif and WD repeat-containing protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IHI
 
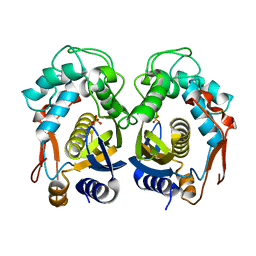 | | Crystal structure of mouse thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2009-07-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
3I7N
 
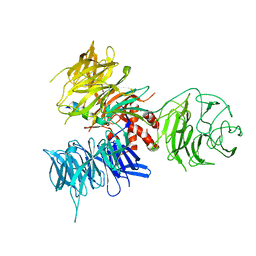 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDTC1 | | Descriptor: | DNA damage-binding protein 1, WD and tetratricopeptide repeats protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1ADO
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ALDOLASE, SULFATE ION | | Authors: | Blom, N.S, Sygusch, J. | | Deposit date: | 1996-12-02 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 4, 1997
|
|
3QWZ
 
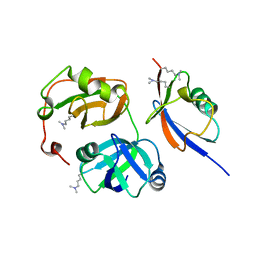 | | Crystal structure of FAF1 UBX-p97N-domain complex | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
