6W6G
 
 | |
1U4J
 
 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
5KVI
 
 | | Crystal structure of monomeric human apoptosis-inducing factor with E413A/R422A/R430A mutations | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Apoptosis-inducing factor 1, mitochondrial, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
5AJA
 
 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
5N2T
 
 | | Thermolysin in complex with inhibitor JC287 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Thermolysin, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N34
 
 | | Thermolysin in complex with inhibitor JC276 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
7ST8
 
 | | Crystal structure of 7H2.2 Fab in complex with SAS1B C-terminal region | | Descriptor: | 7H2.2 Fab Heavy Chain, 7H2.2 Fab Light Chain, Astacin-like metalloendopeptidase | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Monoclonal antibody 7H2.2 binds the C-terminus of the cancer-oocyte antigen SAS1B through the hydrophilic face of a conserved amphipathic helix corresponding to one of only two regions predicted to be ordered
Acta Crystallogr.,Sect.D, 78, 2022
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
5NE7
 
 | |
5L0I
 
 | |
5N6M
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
6W6H
 
 | |
6XUD
 
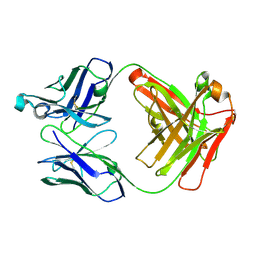 | | Apo Ab 1116NS19.9 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
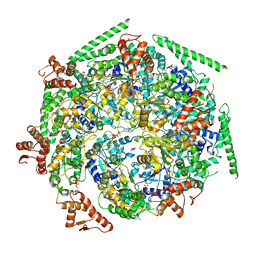
6W6I
 
 | |
5L1G
 
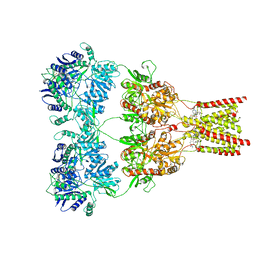 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with GYKI-Br | | Descriptor: | (8R)-5-(4-amino-3-bromophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.507 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
6XUL
 
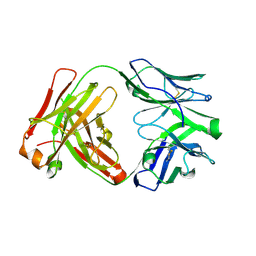 | | Apo Ab 5b1 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6W6E
 
 | |
6W6J
 
 | |
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
5VJB
 
 | |
5AG2
 
 | | SOD-3 azide complex | | Descriptor: | ACETATE ION, AZIDE ION, MALONATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Bonetta, R, Stewart, E.E, Cabelli, D.E, Hunter, T. | | Deposit date: | 2015-01-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Structure of the Caenorhabditis Elegans Manganese Superoxide Dismutase Mnsod-3-Azide Complex.
Protein Sci., 24, 2015
|
|
6XWU
 
 | | Crystal structure of drosophila melanogaster CENP-C cumin domain | | Descriptor: | RE68959p | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
5N9J
 
 | | Core Mediator of transcriptional regulation | | Descriptor: | Mediator Complex Subunit 9, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Nozawa, K, Schneider, T.R, Cramer, P. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Core Mediator structure at 3.4 Angstrom extends model of transcription initiation complex.
Nature, 545, 2017
|
|
