4H2I
 
 | | Human ecto-5'-nucleotidase (CD73): crystal form III (closed) in complex with AMPCP | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
6OLU
 
 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
6O6H
 
 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
6WN0
 
 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
4P7T
 
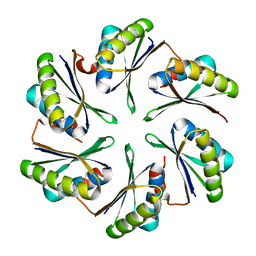 | |
4P7V
 
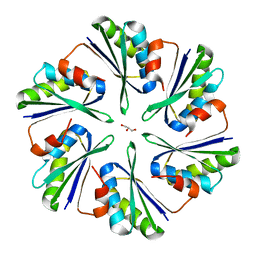 | | Structural insights into higher-order assembly and function of the bacterial microcompartment protein PduA | | Descriptor: | GLYCEROL, Polyhedral bodies | | Authors: | Pang, A, Frank, S, Brown, I.R, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into Higher Order Assembly and Function of the Bacterial Microcompartment Protein PduA.
J.Biol.Chem., 289, 2014
|
|
4S2X
 
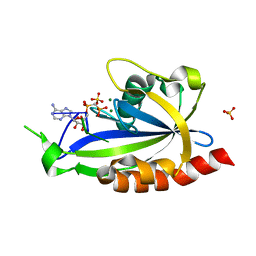 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4Q2A
 
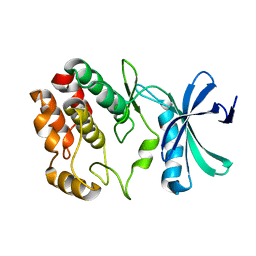 | | WNK1: A chloride sensor via autophosphorylation | | Descriptor: | BROMIDE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Piala, A, Moon, T, Akella, R, He, H, Cobb, M.H, Goldsmith, E. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Chloride Sensing by WNK1 Involves Inhibition of Autophosphorylation.
Sci.Signal., 7, 2014
|
|
6H5H
 
 | |
6HIL
 
 | |
6HIK
 
 | |
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
5V7W
 
 | | Crystal structure of human PARP14 bound to 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one inhibitor | | Descriptor: | 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | Authors: | saikatendu, k.s, Hirozane, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
7AHQ
 
 | | Cryo-EM structure of F-actin stabilized by trans-optoJASP-8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Raunser, S. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-27 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Resolves Molecular Recognition Of An Optojasp Photoswitch Bound To Actin Filaments In Both Switch States.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AHN
 
 | | Cryo-EM structure of F-actin stabilized by cis-optoJASP-8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Raunser, S. | | Deposit date: | 2020-09-24 | | Release date: | 2021-01-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Resolves Molecular Recognition Of An Optojasp Photoswitch Bound To Actin Filaments In Both Switch States.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4AW9
 
 | | Crystal structure of active legumain in complex with YVAD-CMK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, LEGUMAIN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AWB
 
 | | Crystal structure of active legumain in complex with AAN-CMK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEGUMAIN, MERCURY (II) ION, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AWA
 
 | | Crystal structure of active legumain in complex with YVAD-CMK at pH 5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEGUMAIN, SULFATE ION, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BH6
 
 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
4BYY
 
 | | Apo GlxR | | Descriptor: | GLYCEROL, PHOSPHATE ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Pohl, E, Cann, M.J, Townsend, P.D, Money, V.A. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
4BUR
 
 | | Crystal structure of the reduced human Apoptosis inducing factor complexed with NAD | | Descriptor: | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4MFL
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein tcp24, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFK
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA | | Descriptor: | Putative uncharacterized protein tcp24, SULFATE ION, decanoyl-CoA | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFP
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFJ
 
 | | The crystal structure of acyltransferase | | Descriptor: | Putative uncharacterized protein tcp24 | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
