1QVO
 
 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-06-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
1QVP
 
 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
1QVR
 
 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
1QVS
 
 | | Crystal Structure of Haemophilus influenzae H9A mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QVT
 
 | |
1QVU
 
 | |
1QVV
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVW
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
1QVY
 
 | | Crystal structure of RhoGDI K(199,200)R double mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1, SULFATE ION | | Authors: | Czepas, J, Devedjiev, Y, Krowarsh, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-08-29 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The impact of Lys-->Arg surface mutations on the crystallization of the globular domain of RhoGDI.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1QVZ
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QW0
 
 | | Crystal Structure of Haemophilus influenzae N175L mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QW1
 
 | | Solution Structure of the C-Terminal Domain of DtxR residues 110-226 | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
1QW2
 
 | | Crystal Structure of a Protein of Unknown Function TA1206 from Thermoplasma acidophilum | | Descriptor: | conserved hypothetical protein TA1206 | | Authors: | Savchenko, A, Evdokimova, E, Kudrytska, M, Edwards, A.E, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-30 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hypothetical Protein "TA1206" from Thermoplasma acidophilum
To be Published
|
|
1QW4
 
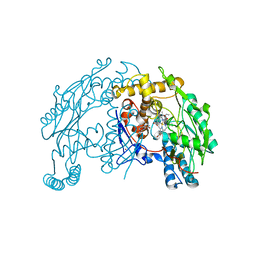 | | Crystal Structure of Murine Inducible Nitric Oxide Synthase Oxygenase Domain in complex with N-omega-propyl-L-arginine. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-PROPYL-L-ARGININE, Nitric oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-08-31 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1QW5
 
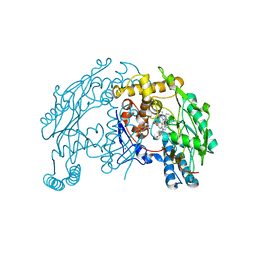 | | Murine inducible nitric oxide synthase oxygenase domain in complex with W1400 inhibitor. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(3-(AMINOMETHYL)BENZYL)ACETAMIDINE, Nitric oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-08-31 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1QW6
 
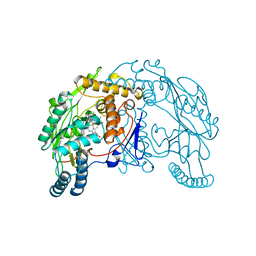 | | Rat neuronal nitric oxide synthase oxygenase domain in complex with N-omega-propyl-L-Arg. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-PROPYL-L-ARGININE, Nitric-oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-09-01 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1QW7
 
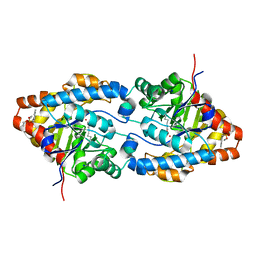 | | Structure of an Engineered Organophosphorous Hydrolase with Increased Activity Toward Hydrolysis of Phosphothiolate Bonds | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, Parathion hydrolase, ... | | Authors: | Mesecar, A.D, Grimsley, J.K, Holton, T, Wild, J.R. | | Deposit date: | 2003-09-01 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mutational studies of organophosphorus hydrolase reveal a cryptic and functional allosteric-binding site.
Arch.Biochem.Biophys., 442, 2005
|
|
1QW8
 
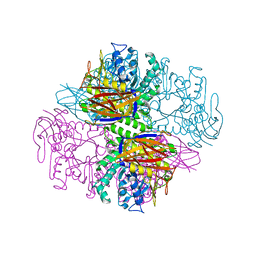 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QW9
 
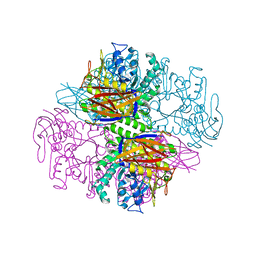 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | Descriptor: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QWA
 
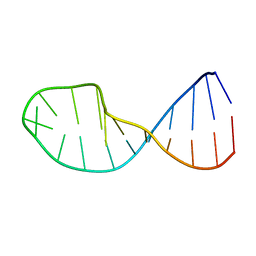 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
1QWB
 
 | |
1QWC
 
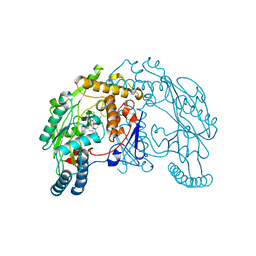 | | Rat neuronal nitric oxide synthase oxygenase domain in complex with W1400 inhibitor. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(3-(AMINOMETHYL)BENZYL)ACETAMIDINE, Nitric-oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-09-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1QWD
 
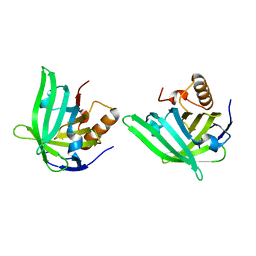 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
1QWE
 
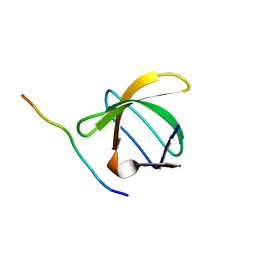 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
