6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
2HSL
 
 | | NMR structure of 13mer duplex DNA containing an abasic site, averaged structure (alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HSS
 
 | | 13mer duplex DNA containg an abasic site with beta anomer, averaged structure | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NYJ
 
 | |
7VCK
 
 | |
6GJU
 
 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
1Z9L
 
 | | 1.7 Angstrom Crystal Structure of the Rat VAP-A MSP Homology Domain | | Descriptor: | Vesicle-associated membrane protein-associated protein A | | Authors: | Kaiser, S.E, Brickner, J.H, Reilein, A.R, Fenn, T.D, Walter, P, Brunger, A.T. | | Deposit date: | 2005-04-03 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of FFAT motif-mediated ER targeting
Structure, 13, 2005
|
|
1Z9O
 
 | | 1.9 Angstrom Crystal Structure of the Rat VAP-A MSP Homology Domain in Complex with the Rat ORP1 FFAT Motif | | Descriptor: | Oxysterol binding protein, Vesicle-associated membrane protein-associated protein A | | Authors: | Kaiser, S.E, Brickner, J.H, Reilein, A.R, Fenn, T.D, Walter, P, Brunger, A.T. | | Deposit date: | 2005-04-03 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of FFAT motif-mediated ER targeting
Structure, 13, 2005
|
|
2LPZ
 
 | | Atomic model of the Type-III Secretion System Needle | | Descriptor: | Protein prgI | | Authors: | Loquet, A, Sgourakis, N.G, Gupta, R, Giller, K, Riedel, D, Goosmann, C, Griesinger, C, Kolbe, M.G, Baker, D, Becker, S, Lange, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of the type III secretion system needle.
Nature, 486, 2012
|
|
2L23
 
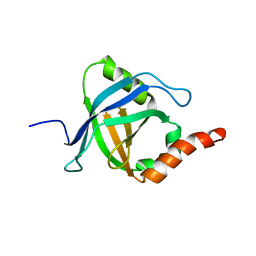 | |
3FFQ
 
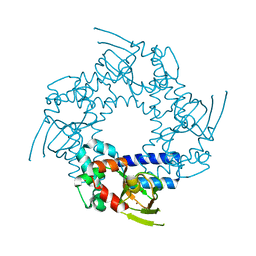 | | HCN2I 443-640 apo-state | | Descriptor: | BROMIDE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Olivier, N.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the structure and conformational movements of proteins with transition metal ion FRET.
Nat.Methods, 6, 2009
|
|
2KNC
 
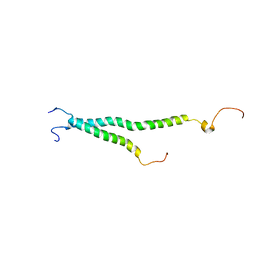 | | Platelet integrin ALFAIIB-BETA3 transmembrane-cytoplasmic heterocomplex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Yang, J, Ma, Y, Page, R.C, Misra, S, Plow, E.F, Qin, J. | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1MAY
 
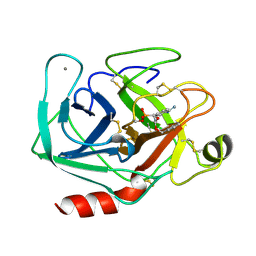 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | Authors: | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | Deposit date: | 1996-02-06 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
2KXI
 
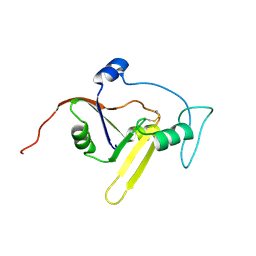 | | Solution NMR structure of the apoform of NarE (NMB1343) | | Descriptor: | Uncharacterized protein | | Authors: | Koehler, C, Carlier, L, Veggi, D, Soriani, M, Pizza, M, Boelens, R, Bonvin, A.M.J.J. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-02 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apoform of NarE (NMB1343)
To be Published
|
|
2KD0
 
 | | NMR solution structure of O64736 protein from Arabidopsis thaliana. Northeast Structural Genomics Consortium MEGA Target AR3445A | | Descriptor: | LRR repeats and ubiquitin-like domain-containing protein At2g30105 | | Authors: | Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of O64736 protein from Arabidopsis thaliana
To be Published
|
|
7A01
 
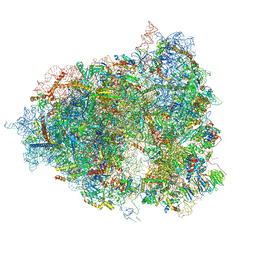 | | The Halastavi arva virus intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES21, ... | | Authors: | Abaeva, I, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T.V, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
7AAZ
 
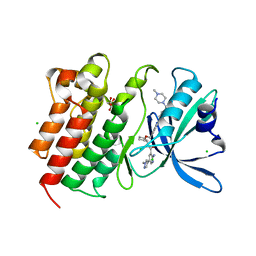 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
3KLR
 
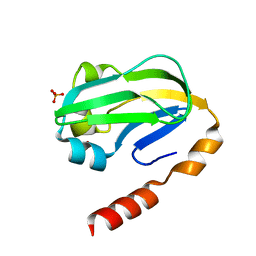 | | Bovine H-protein at 0.88 angstrom resolution | | Descriptor: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | Authors: | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | Deposit date: | 2009-11-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8ALZ
 
 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
3KPW
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 1-Aminoisoquinoline | | Descriptor: | ISOQUINOLIN-1-AMINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
7ADO
 
 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7A2I
 
 | |
3KQP
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Aminoquinoline | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-6-amine | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
5DUI
 
 | |
