2G1J
 
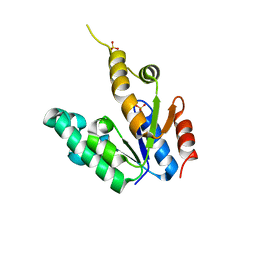 | | Crystal structure of Mycobacterium tuberculosis Shikimate Kinase at 2.0 angstrom resolution | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
4CM5
 
 | |
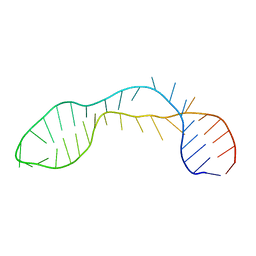
2PCV
 
 | |
6LW8
 
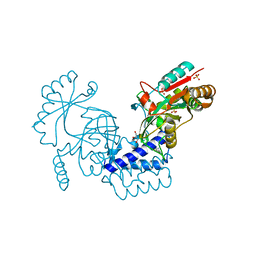 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | (4R)-4-(4-fluorophenyl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, DNA ligase A, GLYCEROL, ... | | Authors: | Ramachandran, R, Afsar, M, Shukla, A. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
3WO4
 
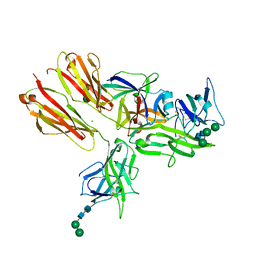 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
4KQ8
 
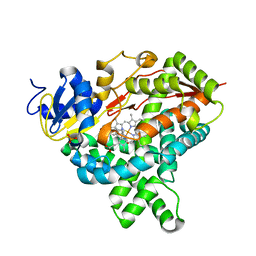 | | Structure of Recombinant Human Cytochrome P450 Aromatase | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Cytochrome P450 19A1, PHOSPHATE ION, ... | | Authors: | Ghosh, D, Di Nardo, G, Griswold, J. | | Deposit date: | 2013-05-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the functional roles of critical residues in human cytochrome p450 aromatase.
Biochemistry, 52, 2013
|
|
4ECS
 
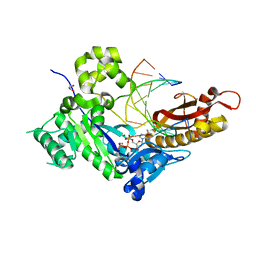 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 80 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ECX
 
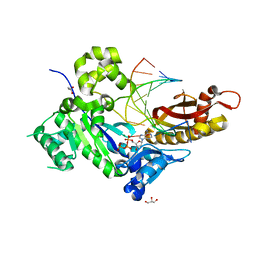 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 300 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ECW
 
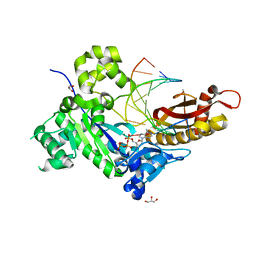 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 250 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4EEY
 
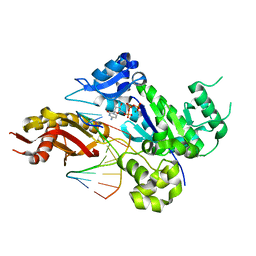 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1RJJ
 
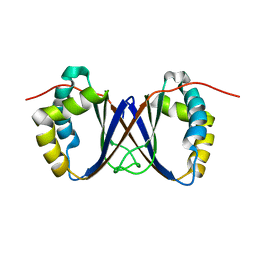 | | Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana | | Descriptor: | expressed protein | | Authors: | Cornilescu, G, Cornilescu, C.C, Zhao, Q, Frederick, R.O, Peterson, F.C, Thao, S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a homodimeric hypothetical protein, at5g22580, a structural genomics target from Arabidopsis thaliana.
J.Biomol.Nmr, 29, 2004
|
|
4FZV
 
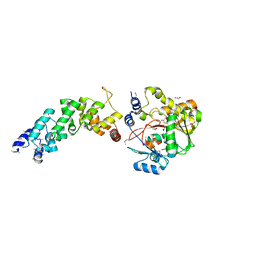 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
1NBO
 
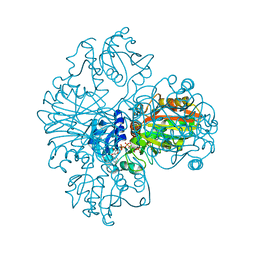 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
2LXT
 
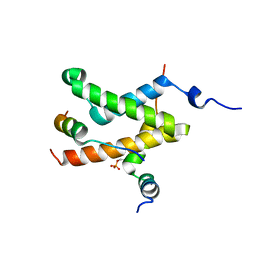 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
7W9K
 
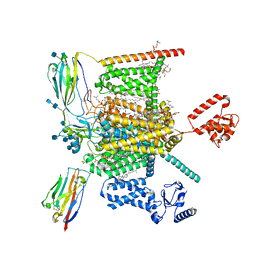 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
6GAA
 
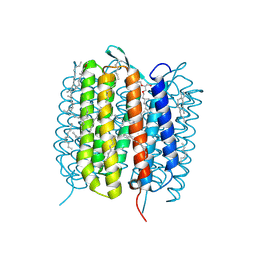 | | BACTERIORHODOPSIN, 430 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6D09
 
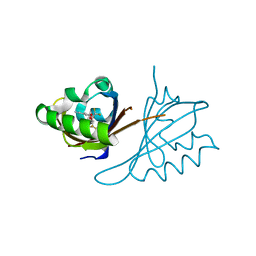 | | Crystal structure of PT2440 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(3R)-4-(difluoromethyl)-2,2-difluoro-3-hydroxy-1,1-dioxo-2,3-dihydro-1H-1-benzothiophen-5-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2018-04-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5L3Z
 
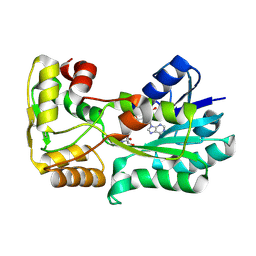 | | polyketide ketoreductase SimC7 - binary complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
6SNO
 
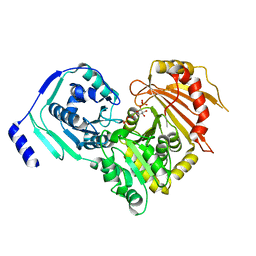 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Phosphoglucomutase-1, ZINC ION | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
6SZG
 
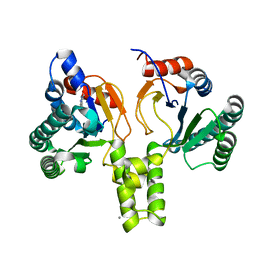 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GR839 and GSK513 | | Descriptor: | (4-chlorophenyl)-[(3~{S})-3-oxidanylpiperidin-1-yl]methanone, 4,5,6,7-tetrahydro-2~{H}-indazole-3-carboxylic acid, CALCIUM ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8CFE
 
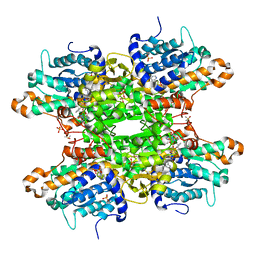 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment B03 | | Descriptor: | 2-[(1R)-1-aminopropyl]phenol, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry B03
To be published
|
|
8CFN
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F03 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F03
To be published
|
|
8CFO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F04 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F04
To be published
|
|
8CFW
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H05 | | Descriptor: | 6-methyl-N~4~-[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H05
To be published
|
|
