7MLL
 
 | |
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
6POJ
 
 | |
7QA9
 
 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7RIK
 
 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
8XEQ
 
 | |
3BBG
 
 | |
5WAH
 
 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | Descriptor: | IgA FC receptor | | Authors: | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
7BWO
 
 | | Consensus chitin binding protein | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
5T0X
 
 | |
7TH8
 
 | | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds | | Descriptor: | Nodule cysteine-rich protein 13 | | Authors: | Buchko, G.W, Zhou, M, Shah, D.M, Velivelli, S.L.S. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds
To Be Published
|
|
6JWE
 
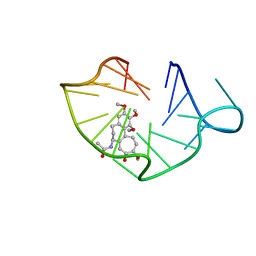 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
7FBR
 
 | |
5LG9
 
 | |
7FBV
 
 | |
7A4L
 
 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | Descriptor: | IRON/SULFUR CLUSTER, PioC | | Authors: | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
1AYG
 
 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
7WCG
 
 | |
1KDL
 
 | |
8T01
 
 | |
6MPP
 
 | |
6KCZ
 
 | |
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | Descriptor: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
1A91
 
 | | SUBUNIT C OF THE F1FO ATP SYNTHASE OF ESCHERICHIA COLI; NMR, 10 STRUCTURES | | Descriptor: | F1FO ATPASE SUBUNIT C | | Authors: | Girvin, M.E, Rastogi, V.K, Abildgaard, F, Markley, J.L, Fillingame, R.H. | | Deposit date: | 1998-04-15 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase.
Biochemistry, 37, 1998
|
|
