7ZVU
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7MXN
 
 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MX7
 
 | | PRMT5:MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXC
 
 | | PRMT5:MEP50 complexed with adenosine | | Descriptor: | ADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXA
 
 | | PRMT5:MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXG
 
 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
6K1S
 
 | | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase (PRMT5) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[7-[(2~{R},3~{R},4~{S},5~{R})-5-[(~{R})-(4-chlorophenyl)-oxidanyl-methyl]-3,4-bis(oxidanyl)oxolan-2-yl]pyrrolo[2,3-d]pyrimidin-4-yl]amino]ethanal, DIMETHYL SULFOXIDE, ... | | Authors: | Tong, S, Lin, H. | | Deposit date: | 2019-05-12 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
1OBH
 
 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A PRE-TRANSFER EDITING SUBSTRATE ANALOGUE IN BOTH SYNTHETIC ACTIVE SITE AND EDITING SITE | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MERCURY (II) ION, NORVALINE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
2ZP1
 
 | |
1OBC
 
 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A POST-TRANSFER EDITING SUBSTRATE ANALOGUE | | Descriptor: | 2'-AMINO-2'-DEOXYADENOSINE, LEUCINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-30 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
6M7A
 
 | |
6M7B
 
 | |
1QTQ
 
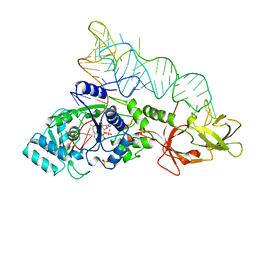 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH TRNA AND AN AMINO ACID ANALOG | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, PROTEIN (GLUTAMINYL-TRNA SYNTHETASE), RNA (TRNA GLN II ), ... | | Authors: | Rath, V.L, Silvian, L.F, Beijer, B, Sproat, B.S, Steitz, T.A. | | Deposit date: | 1998-01-28 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How glutaminyl-tRNA synthetase selects glutamine.
Structure, 6, 1998
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
2RD2
 
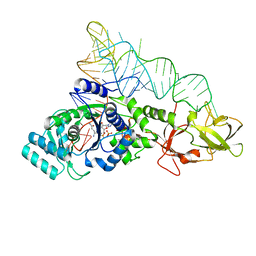 | |
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2RE8
 
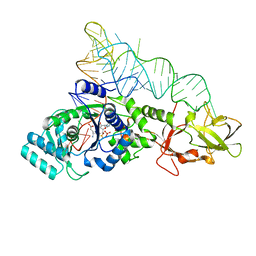 | |
5Z57
 
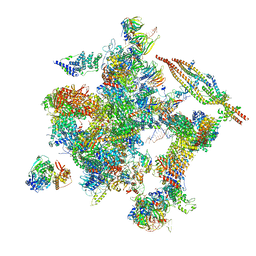 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
1Y8R
 
 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-SUMO1-MG-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | Authors: | Lois, L.M, Lima, C.D. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
1CG3
 
 | | STRUCTURE OF THE MUTANT (R143L) OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH HADACIDIN, GDP, 6-PHOSPHORYL-IMP, AND MG2+ | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Choe, J.Y, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-03-26 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic implications from crystalline complexes of wild-type and mutant adenylosuccinate synthetases from Escherichia coli.
Biochemistry, 38, 1999
|
|
3KYC
 
 | | Human SUMO E1 complex with a SUMO1-AMP mimic | | Descriptor: | 5'-deoxy-5'-(sulfamoylamino)adenosine, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
3KYD
 
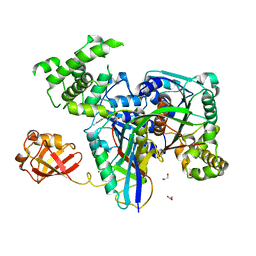 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
8PPK
 
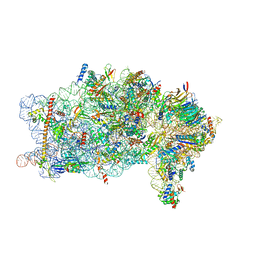 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
