3JTH
 
 | |
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
5UBF
 
 | |
3QP5
 
 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
6UCO
 
 | |
6UCP
 
 | |
1GXR
 
 | | WD40 Region of Human Groucho/TLE1 | | Descriptor: | CALCIUM ION, TRANSDUCIN-LIKE ENHANCER PROTEIN 1 | | Authors: | Pearl, L.H, Roe, S.M, Pickles, L.M. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the C-Terminal Wd40 Repeat Domain of the Human Groucho/Tle1 Transcriptional Corepressor
Structure, 10, 2002
|
|
5UBD
 
 | | Crystal structure of the N-terminal domain (domain 1) of RctB, RctB-1-124-L48M | | Descriptor: | RctB replication initiator protein | | Authors: | Orlova, N, Ivashkiv, O, Waldor, M.K, Jeruzalmi, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The replication initiator of the cholera pathogen's second chromosome shows structural similarity to plasmid initiators.
Nucleic Acids Res., 45, 2017
|
|
8CEZ
 
 | |
3KQK
 
 | |
4RJF
 
 | |
2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
4B09
 
 | | Structure of unphosphorylated BaeR dimer | | Descriptor: | HEXATANTALUM DODECABROMIDE, TRANSCRIPTIONAL REGULATORY PROTEIN BAER | | Authors: | Choudhury, H, Beis, K. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Dimeric Form of the Unphosphorylated Response Regulator Baer.
Protein Sci., 22, 2013
|
|
3QP6
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
1G18
 
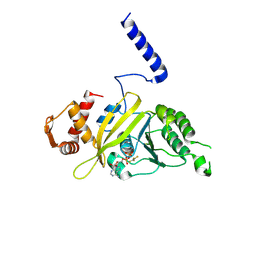 | | RECA-ADP-ALF4 COMPLEX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RECA PROTEIN, TETRAFLUOROALUMINATE ION | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
1G19
 
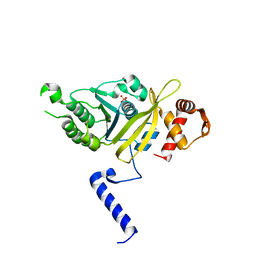 | | STRUCTURE OF RECA PROTEIN | | Descriptor: | PHOSPHATE ION, RECA PROTEIN | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
5UBE
 
 | |
2WAL
 
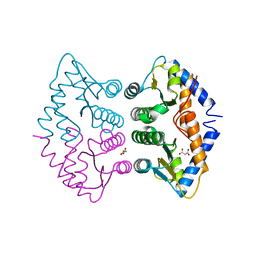 | | Crystal Structure of human GADD45gamma | | Descriptor: | GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE PROTEIN GADD45 GAMMA, MALONIC ACID | | Authors: | Bhattacharya, S, Mueller, J.J, Roske, Y, Turnbull, A.P, Quedenau, C, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Gadd45Gamma
To be Published
|
|
3C28
 
 | |
5CVR
 
 | |
1HST
 
 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
5ZNX
 
 | | Crystal structure of CM14-treated HlyU from Vibrio vulnificus | | Descriptor: | Transcriptional activator | | Authors: | Park, N, Kim, S, Jo, I, Ahn, J, Hong, S, Jeong, S, Baek, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Small-molecule inhibitor of HlyU attenuates virulence of Vibrio species.
Sci Rep, 9, 2019
|
|
6NOG
 
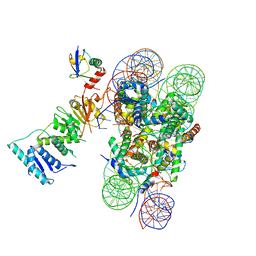 | | Poised-state Dot1L bound to the H2B-Ubiquitinated nucleosome | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
6NJ9
 
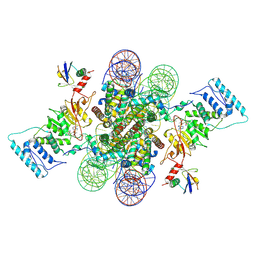 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 2-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
1EO3
 
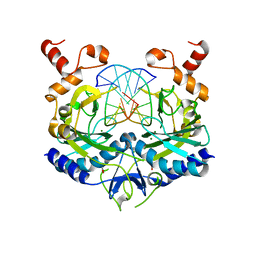 | | INHIBITION OF ECORV ENDONUCLEASE BY DEOXYRIBO-3'-S-PHOSPHOROTHIOLATES: A HIGH RESOLUTION X-RAY CRYSTALLOGRAPHIC STUDY | | Descriptor: | ACETIC ACID, DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
