8IJV
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJW
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
8JBM
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mn2+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JFZ
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E1.Mg2+ state. | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBL
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mg2+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBK
 
 | | Crystal structure of Na+,K+-ATPase in the E1.3Na+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8F2K
 
 | |
3UA9
 
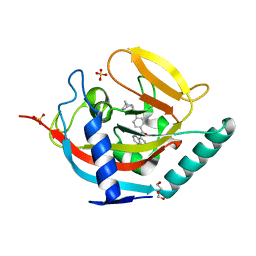 | | Crystal structure of human tankyrase 2 in complex with a selective inhibitor | | Descriptor: | 4-[(3aR,4S,7R,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Narwal, M, Lehtio, L. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of selective inhibition of human tankyrases.
J.Med.Chem., 55, 2012
|
|
3TKD
 
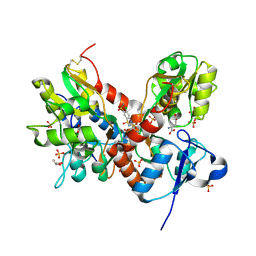 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and cyclothiazide at 1.45 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
6YU4
 
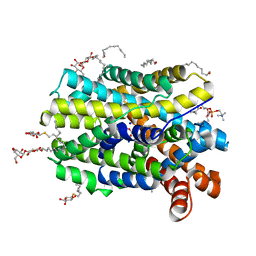 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
7LNU
 
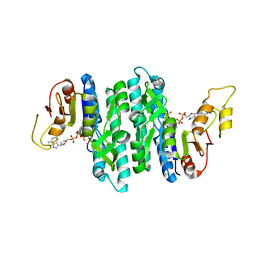 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
8FOB
 
 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Yelshanskaya, M.V, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural mechanism of human oncochannel TRPV6 inhibition by the natural phytoestrogen genistein.
Nat Commun, 14, 2023
|
|
6ZHH
 
 | | Ca2+-ATPase from Listeria Monocytogenes with G4 insertion. | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase, ... | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
6Z2C
 
 | | Engineered lipocalin C3A5 in complex with a transition state analog | | Descriptor: | 1,7,8,9,10,10-hexachloro-4-carboxypentyl-4-aza-tricyclo[5.2.1.0(2,6)]dec-8-ene-3,5-dione, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an engineered lipocalin that catalyzes a Diels-Alder reaction
To be published
|
|
8FQ6
 
 | |
8FR0
 
 | |
8FQ3
 
 | | LBD conformation 2 (LBDconf2) of GluA2 flip Q isoform N619K mutant of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg/N619K) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPC
 
 | | Conformation 1 of the ligand binding domain (LBDconf1) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPH
 
 | | Conformation 2 of the ligand binding domain (LBDconf2) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPL
 
 | | LBD conformation 2 (LBDconf2) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100uM CNQX (Closed-CaNaMg) | | Descriptor: | 6-(aminomethyl)-7-nitro-1,4-dihydroquinoxaline-2,3-dione, CYCLOTHIAZIDE, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQ8
 
 | |
8FQE
 
 | |
8FQD
 
 | |
