2KI5
 
 | | HERPES SIMPLEX TYPE-1 THYMIDINE KINASE IN COMPLEX WITH THE DRUG ACICLOVIR AT 1.9A RESOLUTION | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, PROTEIN (THYMIDINE KINASE), SULFATE ION | | Authors: | Bennett, M.S, Wien, F, Champness, J.N, Batuwangala, T, Rutherford, T, Summers, W.C, Sun, H, Wright, G, Sanderson, M.R. | | Deposit date: | 1999-02-12 | | Release date: | 1999-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir.
FEBS Lett., 443, 1999
|
|
6WPR
 
 | |
1U62
 
 | |
2CK1
 
 | | The structure of oxidised cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin-A from Schistosoma Mansoni: Evidence for Redox- Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
2BOA
 
 | | Human procarboxypeptidase A4. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBOXYPEPTIDASE A4, ... | | Authors: | Garcia-Castellanos, R, Bonet-Figueredo, R, Pallares, I, Ventura, S, Aviles, F.X, Vendrell, J, Gomis-Ruth, F.X. | | Deposit date: | 2005-04-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detailed Molecular Comparison between the Inhibition Mode of A/B-Type Carboxypeptidases in the Zymogen State and by the Endogenous Inhibitor Latexin.
Cell.Mol.Life Sci., 62, 2005
|
|
2CMT
 
 | | The structure of reduced cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin a from Schistosoma Mansoni. Evidence for Redox Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
5X2A
 
 | |
5X26
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2K
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with WZ4003 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Zhu, S.J, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
1XV4
 
 | |
1XV7
 
 | |
5X28
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2C
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(5) | | Descriptor: | 1,2-ETHANEDIOL, 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, ... | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2F
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X27
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
8G2E
 
 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
3LTW
 
 | | The structure of mycobacterium marinum arylamine n-acetyltransferase in complex with hydralazine | | Descriptor: | 1-hydrazinophthalazine, Arylamine N-acetyltransferase Nat, FORMIC ACID | | Authors: | Abuhammad, A.M, Lowe, E.D, Fullam, E, Noble, M, Garman, E.F, Sim, E. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site
Protein Cell, 1, 2010
|
|
7RD0
 
 | |
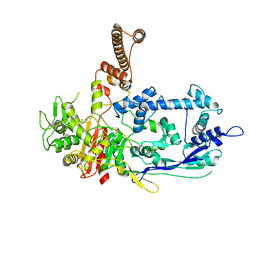
7RCX
 
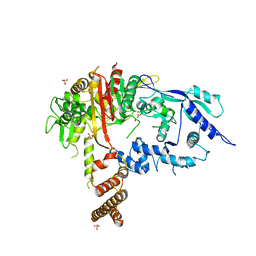 | |
7RCW
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
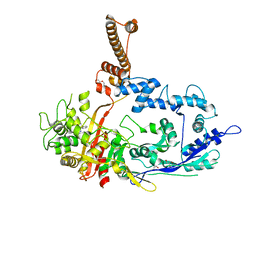
7RCZ
 
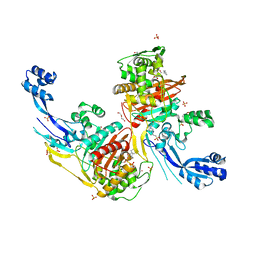 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCY
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
6UDS
 
 | |
6UUV
 
 | |
6UUT
 
 | |
