3UCR
 
 | | Crystal structure of the immunoreceptor TIGIT IgV domain | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Yin, J.P, Stengel, K.F, Rouge, L, Bazan, J.F, Wiesmann, C. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.627 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UPC
 
 | | A general strategy for the generation of human antibody variable domains with increased aggregation resistance | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Dudgeon, K, Rouet, R, Kokmeijer, I, Langley, D.B, Christ, D. | | Deposit date: | 2011-11-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | General strategy for the generation of human antibody variable domains with increased aggregation resistance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UPA
 
 | | A general strategy for the generation of human antibody variable domains with increased aggregation resistance | | Descriptor: | kappa light chain variable domain | | Authors: | Dudgeon, K, Rouet, R, Kokmeijer, I, Langley, D.B, Christ, D. | | Deposit date: | 2011-11-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General strategy for the generation of human antibody variable domains with increased aggregation resistance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UDW
 
 | | Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Rouge, L, Stengel, K.F, Yin, J.P, Bazan, F.J, Wiesmann, C. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WV0
 
 | | O-glycan attached to herpes simplex virus type 1 glycoprotein gB is recognized by the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | Envelope glycoprotein B, N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-galactopyranose, Paired immunoglobulin-like type 2 receptor alpha, ... | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WUZ
 
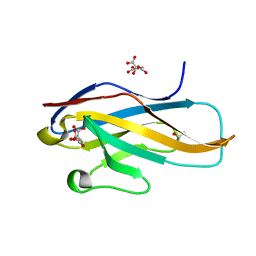 | | Crystal structure of the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8XR5
 
 | | Crystal structure of PD-L1 complexed with small molecule inhibitor X18 | | Descriptor: | (2~{R})-2-[[2-(2,1,3-benzoxadiazol-5-ylmethoxy)-5-chloranyl-4-[[2-fluoranyl-3-[3-[3-(4-oxidanylpiperidin-1-yl)propoxy]phenyl]phenyl]methoxy]phenyl]methylamino]-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Liu, L. | | Deposit date: | 2024-01-06 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel PD-L1 Small-Molecular Inhibitors with Potent In Vivo Anti-tumor Immune Activity.
J.Med.Chem., 67, 2024
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
4QXW
 
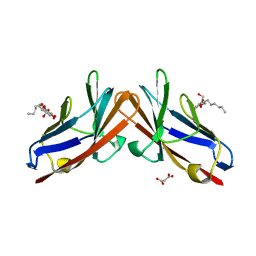 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
8SZY
 
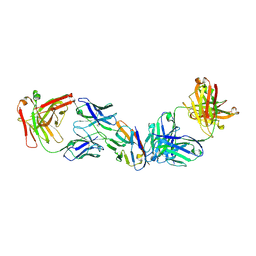 | |
8TBQ
 
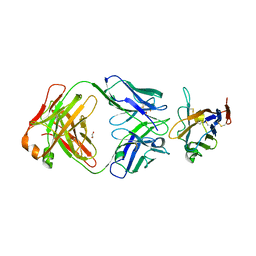 | | The crystal structure of human VISTA extra cellular domain in complex with Fab fragment of pH-selective anti-VISTA antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, SNS-101 Fab heavy chain, ... | | Authors: | van der Horst, E.H, Mukherjee, A, Thisted, T. | | Deposit date: | 2023-06-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | VISTA checkpoint inhibition by pH-selective antibody SNS-101 with optimized safety and pharmacokinetic profiles enhances PD-1 response.
Nat Commun, 15, 2024
|
|
4Y8A
 
 | |
4YIQ
 
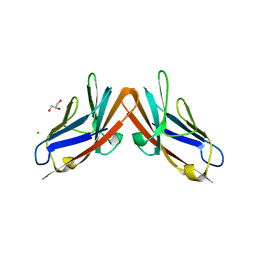 | | Structure of the CEACAM6-CEACAM8 heterodimer | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 6, Carcinoembryonic antigen-related cell adhesion molecule 8, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diverse oligomeric states of CEACAM IgV domains.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WTZ
 
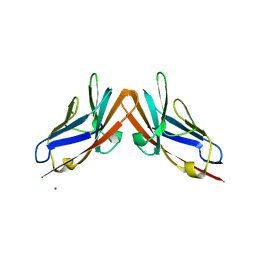 | | Human CEACAM6-CEACAM8 N-domain heterodimer complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 6, Carcinoembryonic antigen-related cell adhesion molecule 8, NICKEL (II) ION | | Authors: | Kirouac, K.N, Prive, G.G. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Human CEACAM6-CEACAM8 N-domain heterodimer complex
To Be Published
|
|
8TBB
 
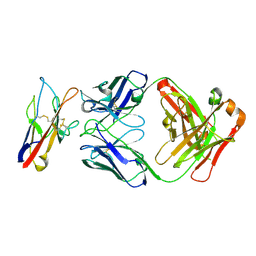 | | F9S, novel TIM-3 targeting antibody, bound to IgV domain of TIM-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F9S Fab heavy chain, F9S Fab light chain, ... | | Authors: | Oganesyan, V, van Dyk, N, Mazor, Y, Yang, C. | | Deposit date: | 2023-06-28 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Generation of AZD7789, a novel PD-1 and TIM-3 targeting bispecific antibody, which binds to a differentiated epitope of TIM-3
To be published
|
|
8TFT
 
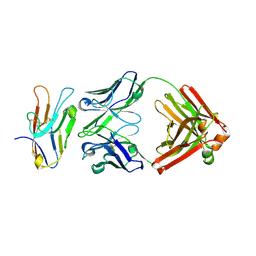 | | Fab of O13-1 human IgG1 antibody bound to IgV domain of human TIM-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O-13-1 IgG1 Fab heavy chain, O-13-1 IgG1 Fab light chain, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Yang, C. | | Deposit date: | 2023-07-11 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation of AZD7789, a novel PD-1 and TIM-3 targeting bispecific antibody, which binds to a differentiated epitope of TIM-3
To be published
|
|
8TE6
 
 | |
8TE5
 
 | |
4Y88
 
 | |
3R0N
 
 | | Crystal Structure of the Immunoglobulin variable domain of Nectin-2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Poliovirus receptor-related protein 2 | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell-cell adhesion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WEU
 
 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ad with llama single domain antibody V3 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AD | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
4WHC
 
 | | Human CEACAM6 N-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 6, ZINC ION | | Authors: | Prive, G.G, Kirouac, K.N. | | Deposit date: | 2014-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Human CEACAM6 N-domain
To Be Published
|
|
3RBG
 
 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
3RNK
 
 | | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain | | Descriptor: | Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain
To be Published
|
|
