4DN4
 
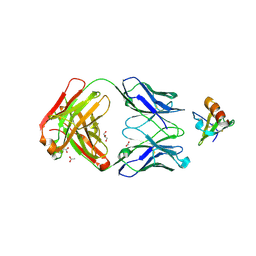 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, 25 STRUCTURES | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
3MU5
 
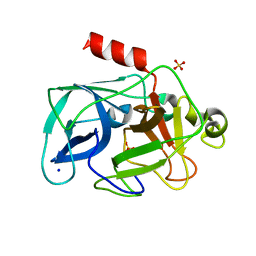 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GQP
 
 | |
4DTE
 
 | |
3MUT
 
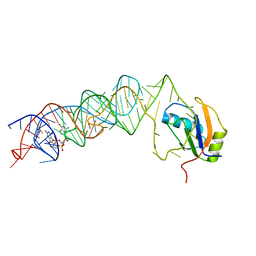 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
4HNQ
 
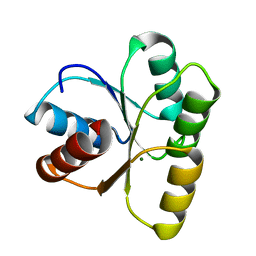 | |
4J14
 
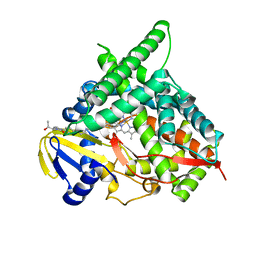 | | Crystal Structure of Human Cytochrome P450 CYP46A1 with Posaconazole Bound | | Descriptor: | Cholesterol 24-hydroxylase, GLYCEROL, POSACONAZOLE, ... | | Authors: | Stout, C.D, Mast, N, Pikuleva, I.A. | | Deposit date: | 2013-01-31 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antifungal Azoles: Structural Insights into Undesired Tight Binding to Cholesterol-Metabolizing CYP46A1.
Mol.Pharmacol., 84, 2013
|
|
4EZP
 
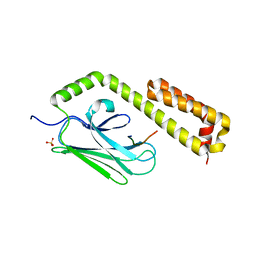 | |
4F01
 
 | |
4AFQ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHYMASE, CITRATE ANION, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4A60
 
 | | Crystal structure of human testis-specific fatty acid binding protein 9 (FABP9) | | Descriptor: | 1,2-ETHANEDIOL, FATTY ACID-BINDING PROTEIN 9 TESTIS LIPID-BINDING PROTEIN, TLBP, ... | | Authors: | Muniz, J.R.C, Kiyani, W, Shrestha, L, Froese, D.S, Krojer, T, Vollmar, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, von Delft, F, Yue, W.W. | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of the Human Fabp9A
To be Published
|
|
4EYN
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
3GXB
 
 | | Crystal structure of VWF A2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Springer, T.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4EZT
 
 | |
4F02
 
 | |
4F0O
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-04 | | Release date: | 2013-05-08 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
3GZP
 
 | |
4F0N
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-04 | | Release date: | 2013-05-08 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
3GZQ
 
 | | HUMAN SOD1 A4V Metal-free Variant | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Galaleldeen, A, Taylor, A.B, Whitson, L.J, Hart, P.J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
3PH5
 
 | |
3PWC
 
 | | Bovine trypsin variant X(tripleGlu217Ile227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3H0B
 
 | | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class | | Descriptor: | 4-[(1S)-1-(3-fluoro-4-methoxyphenyl)-2-(2-methoxy-5-nitrophenyl)ethyl]-1H-imidazol-2-amine, Beta-secretase 1 | | Authors: | Allison, T.J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class: pH dependence on binding activity part 1.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
