2YSH
 
 | | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7 | | Descriptor: | Growth-arrest-specific protein 7 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7
To be Published
|
|
2Q98
 
 | |
3KZ0
 
 | | MCL-1 complex with MCL-1-specific selected peptide | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | Authors: | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
6IJ4
 
 | |
3L02
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92A mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
2QJK
 
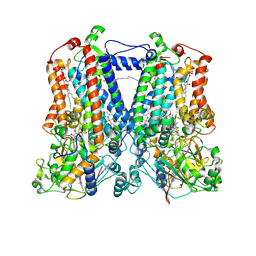 | | Crystal Structure Analysis of mutant rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Esser, L. | | Deposit date: | 2007-07-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides.
J.Biol.Chem., 283, 2008
|
|
8JHZ
 
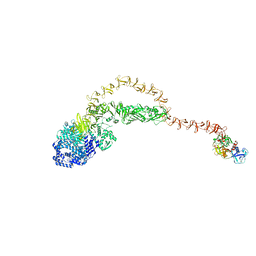 | | Cryo-EM structure of the TcsH-TMPRSS2 complex | | Descriptor: | Hemorrhagic toxin, Transmembrane protease serine 2, ZINC ION | | Authors: | Zhou, R, Liang, T, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
3KO7
 
 | |
1VWR
 
 | |
2QPS
 
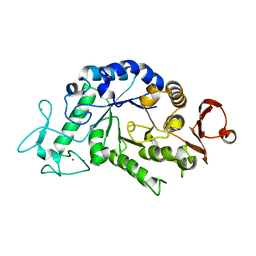 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
1VWP
 
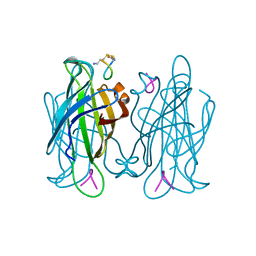 | |
8JI0
 
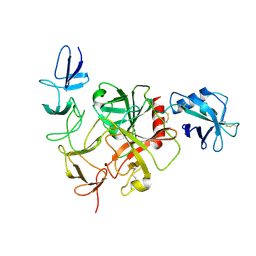 | |
6IMC
 
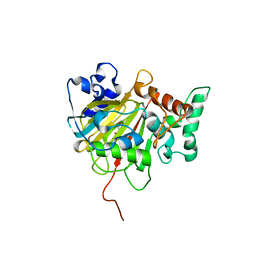 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
2QQU
 
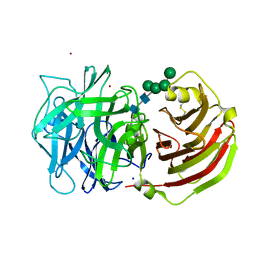 | | Crystal structure of a cell-wall invertase (D239A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
1VWQ
 
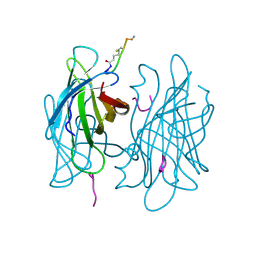 | |
2YSG
 
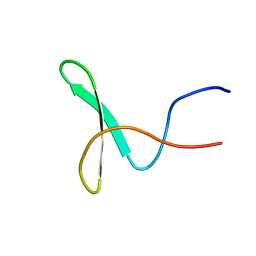 | | Solution structure of the WW domain from the human syntaxin-binding protein 4 | | Descriptor: | Syntaxin-binding protein 4 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human syntaxin-binding protein 4
To be Published
|
|
1VWN
 
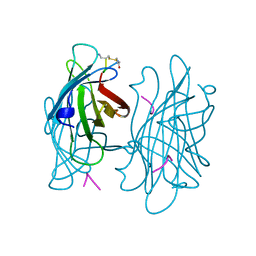 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWM
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.2 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VVD
 
 | |
1VWG
 
 | |
2QMJ
 
 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
1VWJ
 
 | |
2YUD
 
 | | Solution structure of the YTH domain in YTH domain-containing protein 1 (Putative splicing factor YT521) | | Descriptor: | YTH domain-containing protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the YTH domain in YTH domain-containing protein 1 (Putative splicing factor YT521)
To be Published
|
|
8J74
 
 | | Human high-affinity choline transporter CHT1 in the HC-3-bound outward-facing open conformation, dimeric state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), CHOLESTEROL HEMISUCCINATE, HEXADECANE, ... | | Authors: | Gao, Y, Qiu, Y, Zhao, Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
