5JJH
 
 | |
5B68
 
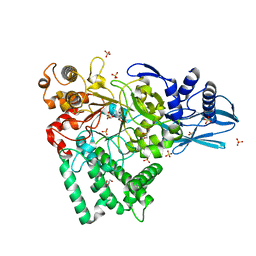 | | Crystal structure of apo amylomaltase from Corynebacterium glutamicum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-alpha-glucanotransferase, GLYCEROL, ... | | Authors: | Joo, S, Kim, S, Kim, K.-J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Amylomaltase from Corynebacterium glutamicum.
J.Agric.Food Chem., 64, 2016
|
|
5F42
 
 | |
5XJH
 
 | |
6IJ6
 
 | |
6IJ3
 
 | |
5YNS
 
 | |
6IJ5
 
 | |
6IJ4
 
 | |
6AG8
 
 | |
6KUS
 
 | |
6KUQ
 
 | |
6KUO
 
 | |
3T57
 
 | |
5Y9X
 
 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and coblat(II) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-08-29 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
5YKA
 
 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with cobalt(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5Y9I
 
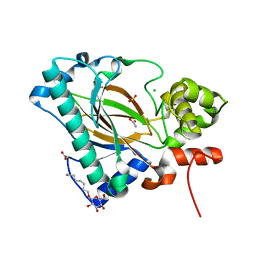 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with Co(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H. | | Deposit date: | 2017-08-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
5Y9Y
 
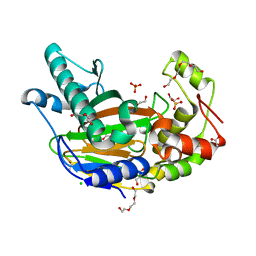 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with succinate and Co(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-08-29 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
5YVZ
 
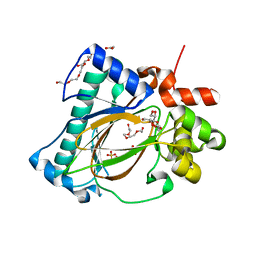 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and Fe(III) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
6A2E
 
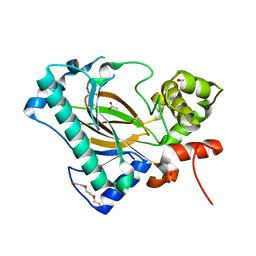 | | Apo structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase | | Descriptor: | ACETATE ION, Kdo hydroxylase, KdoO, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5YW0
 
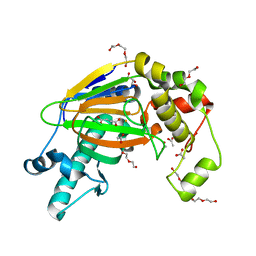 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with succinate and Fe(III) | | Descriptor: | ACETATE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
4J7A
 
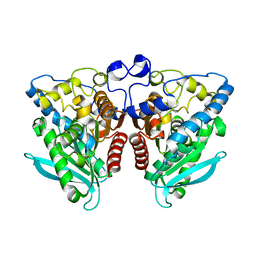 | | Crystal Structure of Est25 - a Bacterial Homolog of Hormone-Sensitive Lipase from a Metagenomic Library | | Descriptor: | Esterase | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Park, K.S, Joo, S.B, Kim, K.K, Kim, D.H. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library
Acta Crystallogr.,Sect.D, 69, 2014
|
|
