5IZ1
 
 | | Physcomitrella patens FBPase | | Descriptor: | fructose-1,6-bisphosphatase | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RQD
 
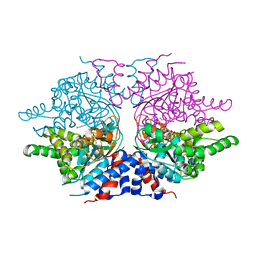 | | deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form with the inhibitor GC7 bound in the active site | | Descriptor: | 1-GUANIDINIUM-7-AMINOHEPTANE, Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-12-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
6PBZ
 
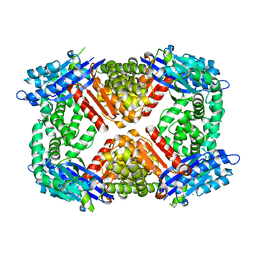 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
1WLU
 
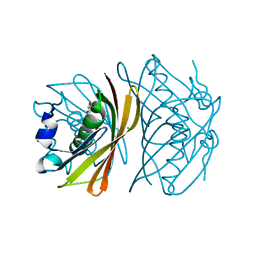 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, phenylacetic acid degradation protein PaaI | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1WN3
 
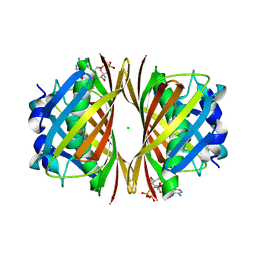 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, HEXANOYL-COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
3SS3
 
 | |
8GA9
 
 | |
8GO6
 
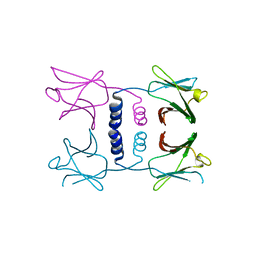 | | Fungal immunomodulatory protein FIP-nha N39A | | Descriptor: | fungal immunomodulatory protein FIP-nha N39A | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
6V27
 
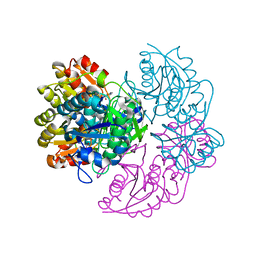 | |
8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
6V26
 
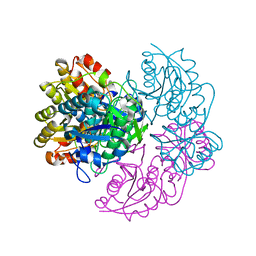 | |
8GO5
 
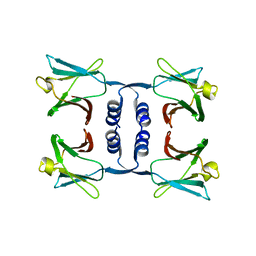 | |
6T65
 
 | |
5K5T
 
 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5JO9
 
 | | Structural characterization of the thermostable Bradyrhizobium japonicum d-sorbitol dehydrogenase | | Descriptor: | PHOSPHATE ION, Ribitol 2-dehydrogenase, sorbitol | | Authors: | Fredslund, F, Otten, H, Navarro Poulsen, J.-C, Lo Leggio, L. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural characterization of the thermostable Bradyrhizobium japonicumD-sorbitol dehydrogenase.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7FIR
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
2VUN
 
 | | The Crystal Structure of Enamidase at 1.9 A Resolution - A new Member of the Amidohydrolase Superfamily | | Descriptor: | CHLORIDE ION, ENAMIDASE, FE (III) ION, ... | | Authors: | Kress, D, Alhapel, A, Pierik, A.J, Essen, L.-O. | | Deposit date: | 2008-05-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Enamidase: A Bifunctional Enzyme of the Nicotinate Catabolism.
J.Mol.Biol., 384, 2008
|
|
7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
8SSF
 
 | | Minimal protein-only/RNA-free Ribonuclease P from Hydrogenobacter thermophilus | | Descriptor: | RNA-free ribonuclease P, SULFATE ION | | Authors: | Mendoza, J, Mallik, L, Wilhelm, C.A, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial RNA-free RNase P: Structural and functional characterization of multiple oligomeric forms of a minimal protein-only ribonuclease P.
J.Biol.Chem., 299, 2023
|
|
7X17
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | Descriptor: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
6ZTX
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6W3Z
 
 | | Crystal Structure of Brugia malayi Deoxyhypusine synthase (DHPS) | | Descriptor: | BMA-DHPS-1, isoform a, CHLORIDE ION, ... | | Authors: | Santiago, A.S, dos Reis, C.V, Ramos, P.Z, Klippel, H.A, Silva, S.F, Zanelli, C.F, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural features and development of an assay platform of the parasite target deoxyhypusine synthase of Brugia malayi and Leishmania major.
Plos Negl Trop Dis, 14, 2020
|
|
6PK3
 
 | |
