3J16
 
 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
2XB4
 
 | | Crystal structures of zinc containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | ADENYLATE KINASE, S,R MESO-TARTARIC ACID, ZINC ION | | Authors: | Mukhopadhyay, A, Kladova, A.V, Gavel, O.Y, Calvete, J.J, Shnyrov, V.L, Moura, I, Moura, J.J.G, Bursakov, S.A, Romao, M.J, Trincao, J. | | Deposit date: | 2010-04-05 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Zinc-, Cobalt-, and Iron-Containing Adenylate Kinase from Desulfovibrio Gigas: A Novel Metal-Containing Adenylate Kinase from Gram-Negative Bacteria.
J.Biol.Inorg.Chem., 16, 2011
|
|
3GMT
 
 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
3J7R
 
 | | Structure of the translating mammalian ribosome-Sec61 complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J7P
 
 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3JBO
 
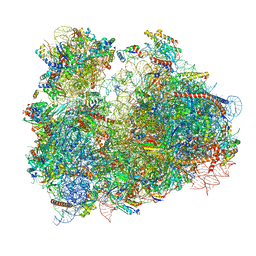 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P/E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3J7A
 
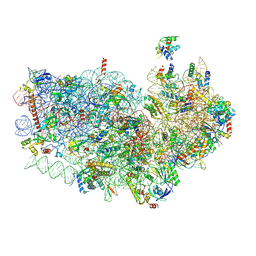 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, small subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
1E4Y
 
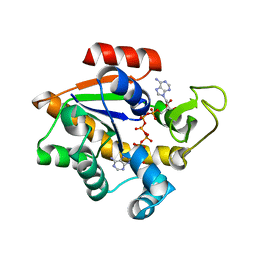 | |
2YZB
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with uric acid (substrate) | | Descriptor: | URIC ACID, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
3JAH
 
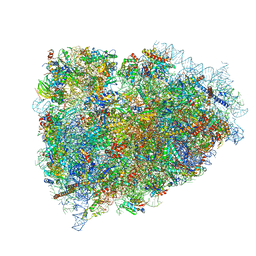 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
2YZC
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with allantoate | | Descriptor: | ALLANTOATE ION, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
3JAN
 
 | |
3J6Y
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 2 degree rotation (Class I) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JAG
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JBP
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
1E4V
 
 | |
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7BZC
 
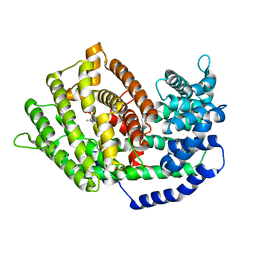 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Z
 
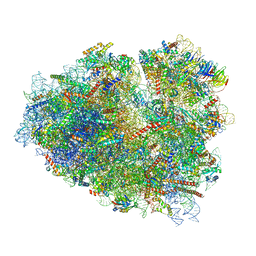 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
6BGV
 
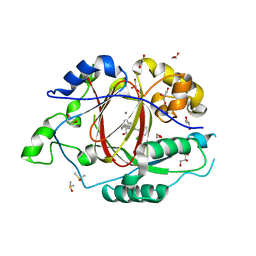 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
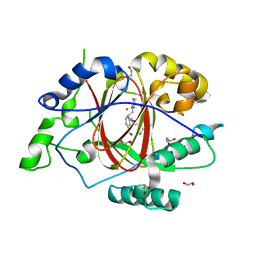 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7NWI
 
 | | Mammalian pre-termination 80S ribosome with Empty-A site bound by Blasticidin S | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA+BlaS, 40S ribosomal protein S11, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
