6ENG
 
 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
8QJ4
 
 | |
8QPF
 
 | |
8QJ3
 
 | |
1CIX
 
 | | THREE-DIMENSIONAL STRUCTURE OF ANTIMICROBIAL PEPTIDE TACHYSTATIN A ISOLATED FROM HORSESHOE CRAB | | Descriptor: | PROTEIN (TACHYSTATIN A) | | Authors: | Fujitani, N, Kawabata, S, Osaki, T, Kumaki, Y, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 1999-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial peptide tachystatin A.
J.Biol.Chem., 277, 2002
|
|
1FQY
 
 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
7Y46
 
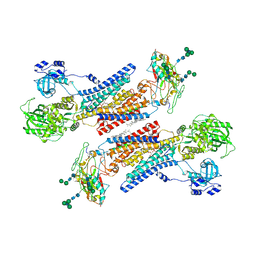 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
4CSK
 
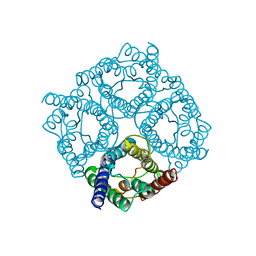 | | human Aquaporin | | Descriptor: | AQUAPORIN-1 | | Authors: | Ruiz-Carrillo, D, To-Yiu-Ying, J, Darwis, D, Soon, C.H, Cornvik, T, Torres, J, Lescar, J. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Crystallization and Preliminary Crystallographic Analysis of Human Aquaporin 1 at a Resolution of 3.28 A.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7W3Y
 
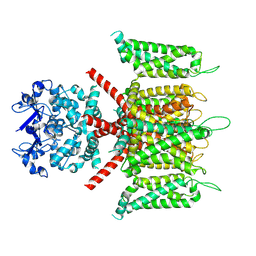 | | CryoEM structure of human Kv4.3 | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
3HZQ
 
 | |
1BKT
 
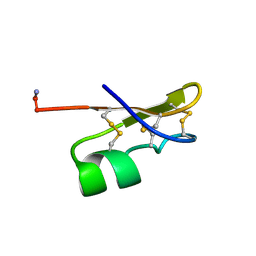 | | BMKTX TOXIN FROM SCORPION BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | BMKTX | | Authors: | Renisio, J.G, Romi-Lebrun, R, Blanc, E, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKTX, a K+ blocker toxin from the Chinese scorpion Buthus Martensi
Proteins, 38, 2000
|
|
7VVH
 
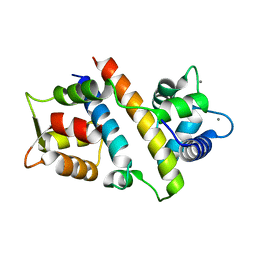 | |
7VVD
 
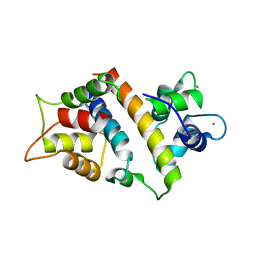 | |
6S5C
 
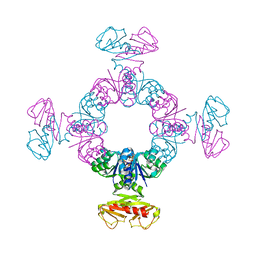 | |
7Y45
 
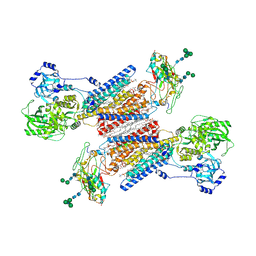 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
1H6I
 
 | |
6E7P
 
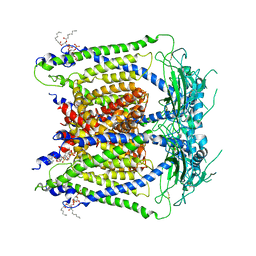 | | cryo-EM structure of human TRPML1 with PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
6E7Y
 
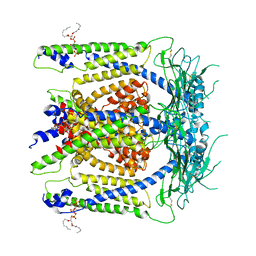 | | cryo-EM structure of human TRPML1 with PI45P2 | | Descriptor: | Mucolipin-1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
6E7Z
 
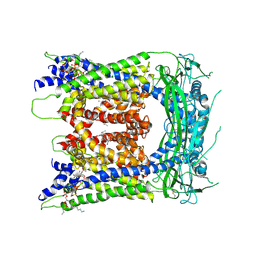 | | cryo-EM structure of human TRPML1 with ML-SA1 and PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
7MGL
 
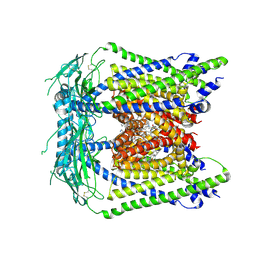 | | Structure of human TRPML1 with ML-SI3 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Mucolipin-1, N-{(1S,2S)-2-[4-(2-methoxyphenyl)piperazin-1-yl]cyclohexyl}benzenesulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic insights into ML-SI3 mediated human TRPML1 inhibition.
Structure, 29, 2021
|
|
6X35
 
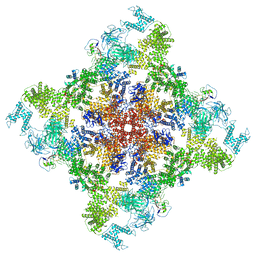 | | Pig R615C RyR1 in complex with CaM, EGTA (class 1, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X33
 
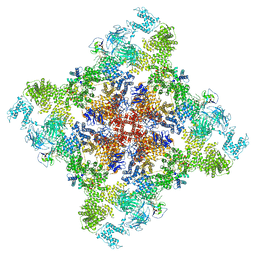 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 3, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6Y94
 
 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6X32
 
 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 1 and 2, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6Y95
 
 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
