5W1U
 
 | | Culex quinquefasciatus carboxylesterase B2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carboxylic ester hydrolase, MALONATE ION | | Authors: | Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Insecticide Sequestering Carboxylesterase from the Disease Vector Culex quinquefasciatus: What Makes an Enzyme a Good Insecticide Sponge?
Biochemistry, 56, 2017
|
|
7Q5T
 
 | | The tandem SH2 domains of SYK with a bound FCER1G diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, High affinity immunoglobulin epsilon receptor subunit gamma, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
8C0G
 
 | | SARS-CoV nsp16-nsp10 complexed with N7-GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 7, SODIUM ION, ... | | Authors: | Ferron, F, Debarnot, C, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Observation of N7-GTP in nsp16-nsp10 Sars-CoV
To Be Published
|
|
5VQA
 
 | | Structure of human TRIP13, ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
8CN7
 
 | | Pa.FabF-C164Q in complex with platencin | | Descriptor: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE | | Authors: | Georgiou, C, Brenk, R. | | Deposit date: | 2023-02-22 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
5W23
 
 | | Crystal Structure of RSV F in complex with 5C4 Fab | | Descriptor: | 5C4 Fab heavy chain, 5C4 Fab light chain, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
7QB7
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345
To Be Published
|
|
8ON5
 
 | |
8C1L
 
 | | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Hepatocyte nuclear factor 4-alpha, Nuclear receptor coactivator 2, ... | | Authors: | Ni, X, Merk, D, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide
To Be Published
|
|
7NPH
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 5-methoxy-1,3-benzoxazole-2-carboxylic acid | | Descriptor: | 5-methoxy-1,3-benzoxazole-2-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase, PHOSPHATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7QC1
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436 | | Descriptor: | 1,2-ETHANEDIOL, Proline--tRNA ligase, [(2~{R},3~{S})-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanylidene-propyl]piperidin-3-yl] ~{N}-[4-[[3-(2,3-dihydro-1~{H}-inden-2-ylcarbamoyl)pyrazin-2-yl]carbamoyl]piperazin-1-yl]sulfonylcarbamate | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436
To Be Published
|
|
7QCQ
 
 | | VHH Z70 in interaction with PHF6 Tau peptide | | Descriptor: | GLYCEROL, Tau 301-312, VHH Z70 | | Authors: | Danis, C, Dupre, E, Zejneli, O, Caillierez, R, Arrial, A, Begard, S, Loyens, A, Mortelecque, J, Cantrelle, F.-X, Hanoulle, X, Rain, J.-C, Colin, M, Buee, L, Landrieu, I. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Tau seeding by targeting Tau nucleation core within neurons with a single domain antibody fragment.
Mol.Ther., 30, 2022
|
|
8ORC
 
 | | Mus Musculus Acetylcholinesterase in complex with AL237 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-(2-methoxyphenyl)thiourea, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.E, Linusson, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Dynamics Determine the Potency and Selectivity of Inhibitors Targeting Disease-Transmitting Mosquitoes.
Acs Infect Dis., 2024
|
|
5KMS
 
 | |
6MJH
 
 | |
8OXG
 
 | | Crystal structure of human methionine aminopeptidase-2 complexed with (3R,4S,5S,6R)-5-methoxy-4-[(2R,3R)-2-methyl-3-(3-methyl-2-buten-1-yl)-2-oxiranyl]-1-oxaspiro[2.5]oct-6-yl N-(trans-4-aminocyclohexyl)carbamate | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, [(1~{R},2~{S},3~{S},4~{R})-2-methoxy-4-methyl-3-[(2~{R},3~{S})-2-methyl-3-(3-methylbut-2-enyl)oxiran-2-yl]-4-oxidanyl-cyclohexyl] ~{N}-(4-azanylcyclohexyl)carbamate | | Authors: | Moss, S, Cornelius, P. | | Deposit date: | 2023-05-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Pharmacological Characterization of SDX-7320/Evexomostat: A Novel Methionine Aminopeptidase Type 2 Inhibitor with Anti-tumor and Anti-metastatic Activity.
Mol.Cancer Ther., 23, 2024
|
|
8BZF
 
 | | FC-J acetonide stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin J-acetonide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
5CX9
 
 | | Crystal structure of CK2alpha with (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoate bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6MJP
 
 | | LptB(E163Q)FGC from Vibrio cholerae | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-cyclohexylhexyl beta-D-glucopyranoside, ABC transporter ATP-binding protein, ... | | Authors: | Owens, T.W, Kahne, D, Kruse, A.C. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
7NPJ
 
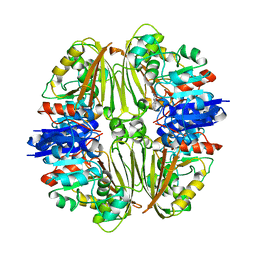 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 6-phenoxy-3-pyridinamine | | Descriptor: | 6-phenoxy-3-pyridinamine, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-27 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7UFV
 
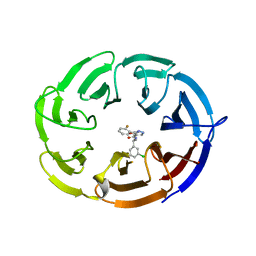 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
5KQS
 
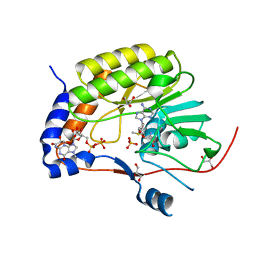 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine and 7-methyl-guanosine-5'-diphosphate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, GLYCEROL, ... | | Authors: | Coloma, J, Jain, R, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
8C08
 
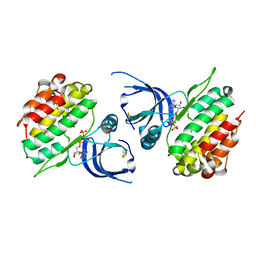 | | Crystal structure of JAK2 JH2-K539L | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
8C14
 
 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chloranyl-3-cyano-phenyl)-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Selective Aurora A-TPX2 Interaction Inhibitors Have In Vivo Efficacy as Targeted Antimitotic Agents.
J.Med.Chem., 67, 2024
|
|
5KR0
 
 | | Protease E35D-APV | | Descriptor: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
