8EKC
 
 | |
6RHW
 
 | | Crystal structure of human CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH | | Descriptor: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | Authors: | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4P5N
 
 | | Structure of CNAG_02591 from Cryptococcus neoformans | | Descriptor: | CADMIUM ION, GLYCEROL, Hypothetical protein CNAG_02591, ... | | Authors: | Ramagopal, U.A, McClelland, E.E, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals.
Plos Pathog., 12, 2016
|
|
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
8UX5
 
 | |
6RZL
 
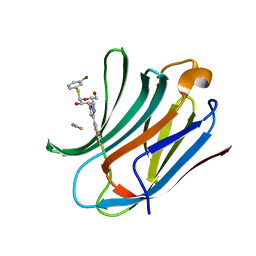 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:4(Bromine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-bromophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.045 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
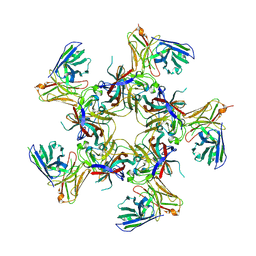
8U5L
 
 | |
7KDT
 
 | |
6USX
 
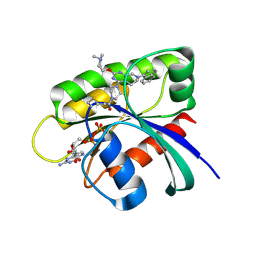 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | 1-{4-[2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
6UT0
 
 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
6UXF
 
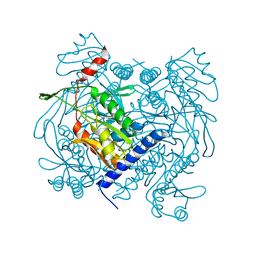 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8HF0
 
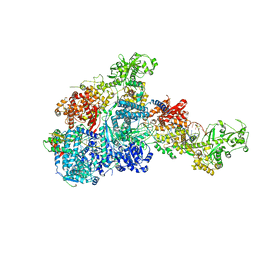 | | DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state | | Descriptor: | Dicer-2, isoform A, LD06392p, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers.
Nat Commun, 14, 2023
|
|
6UVM
 
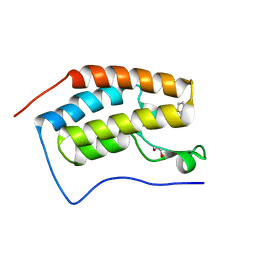 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
6RZH
 
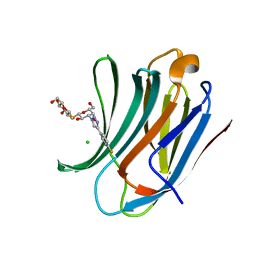 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
8UL7
 
 | | The structure of NanH in complex with Neu5Ac | | Descriptor: | 1,2-ETHANEDIOL, N-acetyl-alpha-neuraminic acid, Sialidase | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
8URL
 
 | |
8UVV
 
 | | The NanJ sialidase catalytic domain in complex with Neu5Ac | | Descriptor: | CHLORIDE ION, Exo-alpha-sialidase NanJ, N-acetyl-alpha-neuraminic acid | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-11-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
8UB5
 
 | | The Apo NanH structure from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Sialidase | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
8ULE
 
 | | The structure of NanH in complex with Neu5,9Ac | | Descriptor: | 1,2-ETHANEDIOL, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, Sialidase | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
8UM0
 
 | | The structure of NanH in complex with Neu5,7,9Ac(2,6)-LAcNAc | | Descriptor: | 1,2-ETHANEDIOL, 5-acetamido-7,9-di-O-acetyl-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, Sialidase, ... | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-10-17 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
8UW1
 
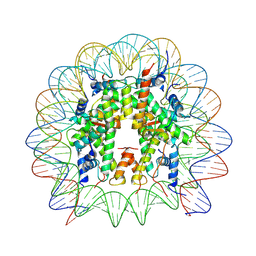 | | Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome | | Descriptor: | DNA (146-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A, ... | | Authors: | Gretarsson, K, Abini-Agbomson, S, Armache, K.-J, Lu, C. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Sci Adv, 10, 2024
|
|
6V4L
 
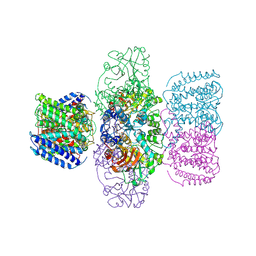 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
8EXH
 
 | | Agrobacterium tumefaciens Tpilus | | Descriptor: | (14S,17R)-20-amino-17-hydroxy-11,17-dioxo-12,16,18-trioxa-17lambda~5~-phosphaicosan-14-yl tetradecanoate, Protein virB2 | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery
Nat Commun, 14, 2023
|
|
6UXG
 
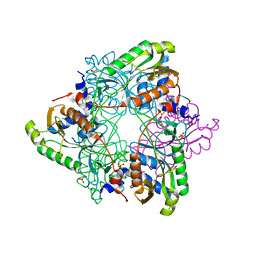 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8HI9
 
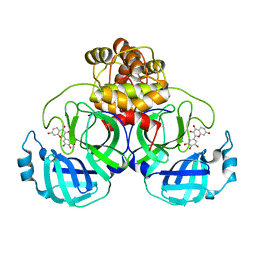 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
