7BG3
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5MV0
 
 | | Structure of an N-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein, PHOSPHATE ION | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
6K3L
 
 | | Crystal structure of CX-4945 bound Cka1 from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CMGC/CK2 protein kinase, SULFATE ION | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
5BQB
 
 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form III | | Descriptor: | CHLORIDE ION, CITRIC ACID, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
5NAN
 
 | | Crystal Structure of human IL-17AF in complex with human IL-17RA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Rondeau, J.-M, Goepfert, A. | | Deposit date: | 2017-02-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The human IL-17A/F heterodimer: a two-faced cytokine with unique receptor recognition properties.
Sci Rep, 7, 2017
|
|
8CHT
 
 | |
8P81
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
7NSV
 
 | | 14-3-3 sigma with p65 (RelA) binding site pS45 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, GLYCEROL, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NQP
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-02 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6M91
 
 | | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-103094 ternary complex | | Descriptor: | 3-({4-[(2,6-dichlorophenyl)sulfanyl]-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carbonyl}amino)benzoic acid, CHLORIDE ION, Catenin beta-1, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
6M93
 
 | | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-1933 ternary complex | | Descriptor: | 2-oxo-N-[3-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide, Catenin beta-1, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Lee, S.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
6M92
 
 | | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-2663 ternary complex | | Descriptor: | 3-{[2-oxo-4-phenoxy-6-(trifluoromethyl)-1,2-dihydropyridine-3-carbonyl]amino}benzoic acid, Catenin beta-1, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J, Lee, S.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
6VCD
 
 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | Authors: | Wang, H, Shi, H, Zheng, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
3R7W
 
 | | Crystal Structure of Gtr1p-Gtr2p complex | | Descriptor: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | Authors: | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-24 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
1UZG
 
 | | CRYSTAL STRUCTURE OF THE DENGUE TYPE 3 VIRUS ENVELOPE PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAJOR ENVELOPE PROTEIN E, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modis, Y, Harrison, S.C. | | Deposit date: | 2004-03-11 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein
J.Virol., 79, 2005
|
|
6ZV1
 
 | | TFIIS N-terminal domain (TND) from human IWS1 | | Descriptor: | Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV2
 
 | | TFIIS N-terminal domain (TND) from human PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV4
 
 | | Human TFIIS N-terminal domain in complex with IWS1 | | Descriptor: | Transcription elongation factor A protein 1,Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV0
 
 | | TFIIS N-terminal domain (TND) from human LEDGF/p75 | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
7NMH
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-070 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) methanesulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
5XQM
 
 | |
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
1ZQ3
 
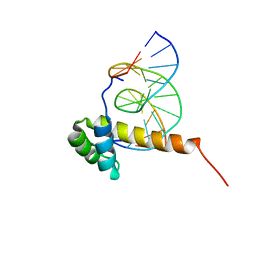 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
7ZMU
 
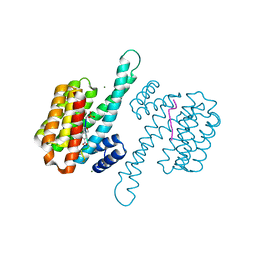 | |
