3ZI3
 
 | | Crystal structure of the B24His-insulin - human analogue | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Zakova, L, Kletvikova, E, Veverka, V, Lepsik, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2013-01-02 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Integrity of the B24 Site in Human Insulin is Important for Hormone Functionality
J.Biol.Chem., 288, 2013
|
|
1KY6
 
 | |
1YZ5
 
 | |
3BF6
 
 | | Thrombin:suramin complex | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, PHE-PRO-ARG, Thrombin, ... | | Authors: | Lima, L.M.T.R, Polikarpov, I, Monteiro, R.Q. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-25 | | Last modified: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases.
Biochim.Biophys.Acta, 1794, 2009
|
|
3B7G
 
 | | Human DEAD-box RNA helicase DDX20, Conserved domain I (DEAD) in complex with AMPPNP (Adenosine-(Beta,gamma)-imidotriphosphate) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
1JLT
 
 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ZFT
 
 | |
1JAU
 
 | |
4IDW
 
 | | Polycrystalline T6 Bovine Insulin: Anisotropic Lattice Evolution and Novel Structure Refinement Strategy | | Descriptor: | Insulin A chain, Insulin B chain, ZINC ION | | Authors: | Margiolaki, I, Giannopoulou, A.E, Wright, J.P, Knight, L, Norrman, M, Schluckebier, G, Fitch, A, Von Dreele, R.B. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-05 | | Method: | POWDER DIFFRACTION | | Cite: | High-resolution powder X-ray data reveal the T6 hexameric form of bovine insulin
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5WRH
 
 | |
5AGA
 
 | | Crystal structure of the Helicase domain of human DNA polymerase theta in complex with AMPPNP | | Descriptor: | CITRATE ANION, DNA POLYMERASE THETA, MAGNESIUM ION, ... | | Authors: | Newman, J.A, Cooper, C.D.O, Aitkenhead, H, Pinkas, D.M, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-25 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway.
Structure, 23, 2015
|
|
5AI7
 
 | | ParM doublet model | | Descriptor: | PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles
Nature, 523, 2015
|
|
3ZEZ
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
1XMZ
 
 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
2ZJA
 
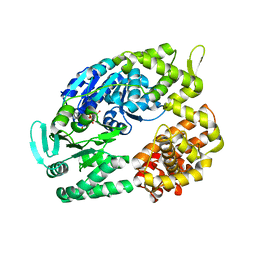 | | Archaeal DNA helicase Hjm complexed with AMPPCP in form 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
4PJ3
 
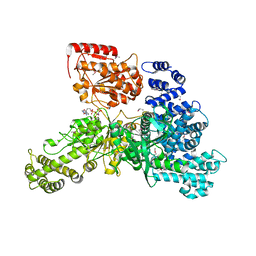 | | Structural insight into the function and evolution of the spliceosomal helicase Aquarius, Structure of Aquarius in complex with AMPPNP | | Descriptor: | Intron-binding protein aquarius, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | De, I, Bessonov, S, Hofele, R, dos Santos, K.F, Will, C.L, Urlaub, H, Luhrmann, R, Pena, V. | | Deposit date: | 2014-05-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5GQO
 
 | |
1L6F
 
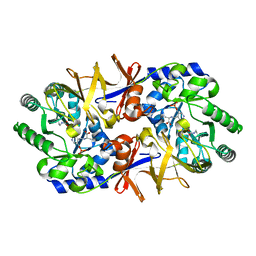 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-L-alanine | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-09 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
3HSG
 
 | | Crystal structure of E. coli HPPK(Y53A) in complex with MgAMPCPP | | Descriptor: | ACETATE ION, CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
3ZF2
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
5XNA
 
 | | Crystal structure of a secretary abundant heat soluble (SAHS) protein from Ramazzottius varieornatus (from dimer sample) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Fukuda, Y, Miura, Y, Mizohata, E, Inoue, T. | | Deposit date: | 2017-05-19 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into a secretory abundant heat-soluble protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus
FEBS Lett., 591, 2017
|
|
1L9N
 
 | |
5SYC
 
 | |
5XVO
 
 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing DNA sampling and half-integration states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
2ZVM
 
 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
