6A5I
 
 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
6P49
 
 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P46
 
 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P48
 
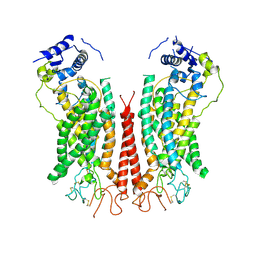 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
4KVT
 
 | | Crystal structure of a 6-helix coiled coil CC-Hex-L24C | | Descriptor: | 6-helix coiled coil CC-Hex-L24C peptide | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
3JXO
 
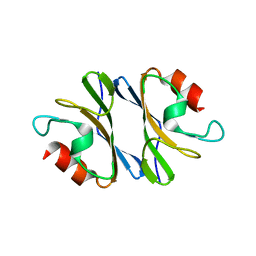 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | TrkA-N domain protein | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-09-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
4AV6
 
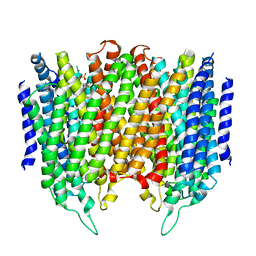 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase at 4 A in complex with phosphate and magnesium | | Descriptor: | K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kajander, T, Kellosalo, J, Kogan, K, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
2MSF
 
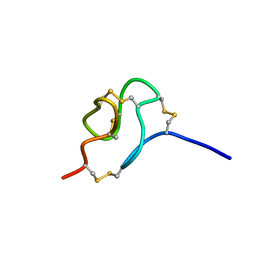 | | NMR SOLUTION STRUCTURE OF SCORPION VENOM TOXIN Ts11 (TsPep1) FROM Tityus serrulatus | | Descriptor: | Peptide TsPep1 | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Cremonez, C.M, Peigneur, S, Cassoli, J.S, Dutra, A.A.A, Waelkens, E, Pimenta, A.M.C, De Lima, M.H, Tytgat, J, Arantes, E.C. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Toxins (Basel), 8, 2016
|
|
2E6W
 
 | | Solution structure and calcium binding properties of EF-hands 3 and 4 of calsenilin | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Yu, L, Sun, C, Mendoza, R, Hebert, E, Pereda-Lopez, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of EF-hands 3 and 4 of calsenilin.
Protein Sci., 16, 2007
|
|
3R48
 
 | | Crystal structure of a hetero-hexamer coiled coil | | Descriptor: | GLYCEROL, coiled coil helix W22-L24H, coiled coil helix Y15-L24D | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0011 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
7L83
 
 | | NMR solution structure of Nav1.5 DIV S3b-S4a paddle motif in DPC micelle | | Descriptor: | Sodium channel protein type 5 subunit alpha | | Authors: | Hussein, A.K, Bhuiyan, M.H, Arshava, B, Zhuang, J, Poget, S.F. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and analysis of isolated S3b-S4a motif of repeat IV of the human cardiac sodium channel
Biorxiv, 2021
|
|
6XQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UAA-mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1RE9
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450-CAM WITH A FLUORESCENT PROBE D-8-AD (ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-06 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires.
J.Mol.Biol., 344, 2004
|
|
4EKN
 
 | |
1SCY
 
 | |
5LIA
 
 | | Crystal structure of murine autotaxin in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turnbull, A.P, Shah, P, Cheasty, A, Raynham, T, Pang, L, Owen, P. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of potent inhibitors of the lysophospholipase autotaxin.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
1C55
 
 | |
1C56
 
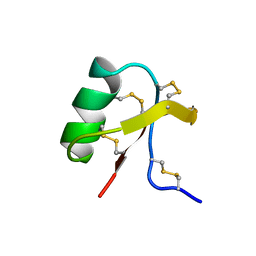 | |
2MCR
 
 | | Solution structure of ShK-like immunomodulatory peptide from Brugia malayi (filarial worm) | | Descriptor: | Probable zinc metalloproteinase, putative | | Authors: | Chhabra, S, Swarbrick, J.D, Pennington, M.W, Chang, S.C, Norton, R.S. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
2MD0
 
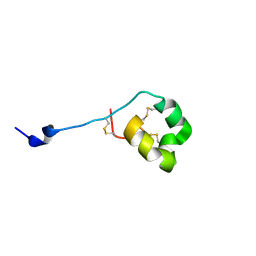 | | Solution structure of ShK-like immunomodulatory peptide from Ancylostoma caninum (hookworm) | | Descriptor: | AcK1 | | Authors: | Chhabra, S, Swarbrick, J.D, Mohanty, B, Chang, S.C, Chandy, G.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
2OZL
 
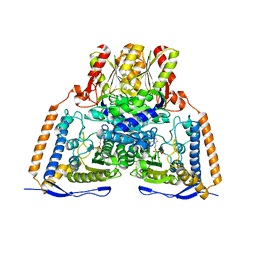 | | Human pyruvate dehydrogenase S264E variant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component alpha subunit, ... | | Authors: | Ciszak, E.M, Dominiak, P.M, Patel, M.S, Korotchkina, L.G. | | Deposit date: | 2007-02-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of Serine 264 Impedes Active Site Accessibility in the E1 Component of the Human Pyruvate Dehydrogenase Multienzyme Complex
Biochemistry, 46, 2007
|
|
3POT
 
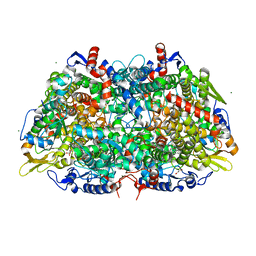 | |
5X24
 
 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5XXI
 
 | | Crystal structure of CYP2C9 in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
