1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLA
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLE
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1IKV
 
 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1HQU
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | Authors: | Hsiou, Y, Ding, J, Arnold, E. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1JLC
 
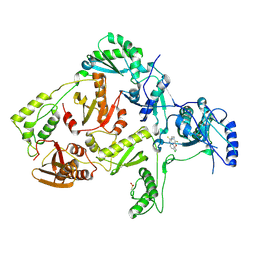 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
2BX3
 
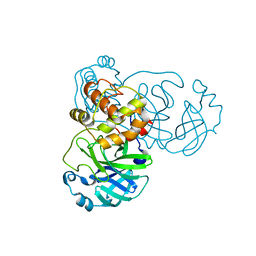 | |
2BX4
 
 | |
3DRR
 
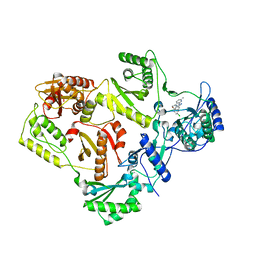 | | HIV reverse transcriptase Y181C mutant in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DLG
 
 | | Crystal structure of hiv-1 reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, P51 RT, PHOSPHATE ION, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DYA
 
 | | HIV-1 RT with non-nucleoside inhibitor annulated Pyrazole 1 | | Descriptor: | 3-[6-bromo-2-fluoro-3-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-chlorobenzonitrile, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
3DOL
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DRS
 
 | | HIV reverse transcriptase K103N mutant in complex with inhibitor R8D | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase/ribonuclease H, p66 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DOK
 
 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | Descriptor: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DRP
 
 | | HIV reverse transcriptase in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3E01
 
 | | HIV-RT with non-nucleoside inhibitor annulated pyrazole 2 | | Descriptor: | 3-[2-bromo-4-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-methylbenzonitrile, Gag-Pol polyprotein | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
3DLE
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
2R2S
 
 | | Co(III)bleomycinB2 bound to d(ATTAGTTATAACTAAT) complexed with MMLV RT catalytic fragment | | Descriptor: | BLEOMYCIN B2, COBALT (III) ION, DNA (5'-D(*DAP*DTP*DTP*DAP*DGP*DTP*DT)-3'), ... | | Authors: | Goodwin, K.D, Lewis, M.A, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2007-08-27 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R2T
 
 | | d(ATTTAGTTAACTAAAT) complexed with MMLV RT catalytic fragment | | Descriptor: | DNA (5'-D(*DAP*DTP*DTP*DTP*DAP*DGP*DTP*DT)-3'), DNA (5'-D(P*DAP*DAP*DCP*DTP*DAP*DAP*DAP*DT)-3'), Reverse transcriptase | | Authors: | Goodwin, K.D, Lewis, M.A, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2007-08-27 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6EXX
 
 | | Crystal Structure of Pes4 RRM4 | | Descriptor: | Protein PES4 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Pes4 RRM4 at 1.1 Angstroms resolution
To Be Published
|
|
6EZ7
 
 | | Pes4 RRM3 Structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein PES4 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pes4 RRM3 Structure
To Be Published
|
|
7L7I
 
 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
1RTD
 
 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | Descriptor: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | Authors: | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
