3I6U
 
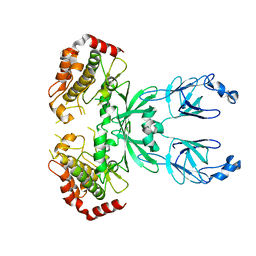 | |
3FAN
 
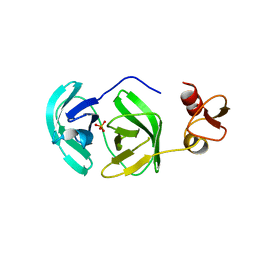 | | Crystal structure of chymotrypsin-like protease/proteinase (3CLSP/Nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3F8S
 
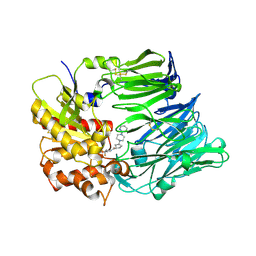 | | Crystal structure of dipeptidyl peptidase IV in complex with inhibitor | | Descriptor: | 2-(4-{(3S,5S)-5-[(3,3-difluoropyrrolidin-1-yl)carbonyl]pyrrolidin-3-yl}piperazin-1-yl)pyrimidine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ammirati, M.J, Liu, S, Piotrowski, D.W. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | (3,3-Difluoro-pyrrolidin-1-yl)-[(2S,4S)-(4-(4-pyrimidin-2-yl-piperazin-1-yl)-pyrrolidin-2-yl]-methanone: a potent, selective, orally active dipeptidyl peptidase IV inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FO6
 
 | | Crystal structure of guanine riboswitch bound to 6-O-methylguanine | | Descriptor: | 6-O-methylguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2008-12-28 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs.
Structure, 17, 2009
|
|
3FIA
 
 | | Crystal structure of the EH 1 domain from human intersectin-1 protein. Northeast Structural Genomics Consortium target HR3646e. | | Descriptor: | CALCIUM ION, Intersectin-1, SULFATE ION | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Devices, C, Zhao, L, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the EH 1 domain from human intersectin-1 protein.
To be Published
|
|
3F71
 
 | |
3F7Q
 
 | |
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
3FAO
 
 | | Crystal structure of S118A mutant 3CLSP of PRRSV | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3EWH
 
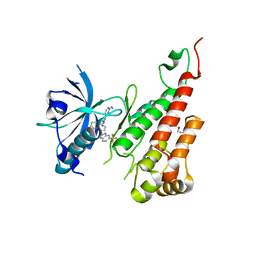 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridyl-pyrimidine benzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-6-(trifluoromethyl)-1H-benzimidazol-2-amine, vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FDD
 
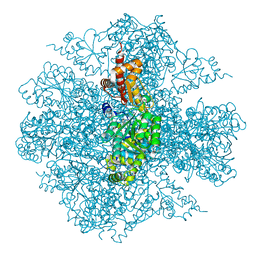 | | The Crystal Structure of the Pseudomonas dacunhae Aspartate-Beta-Decarboxylase Reveals a Novel Oligomeric Assembly for a Pyridoxal-5-Phosphate Dependent Enzyme | | Descriptor: | ACETATE ION, CHLORIDE ION, L-aspartate-beta-decarboxylase, ... | | Authors: | Lima, S, Sundararaju, B, Huang, C, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the Pseudomonas dacunhae aspartate-beta-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme.
J.Mol.Biol., 388, 2009
|
|
3GOT
 
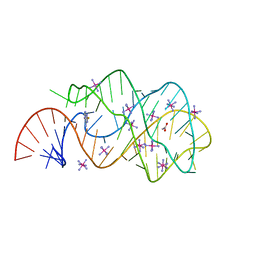 | | Guanine riboswitch C74U mutant bound to 2-fluoroadenine. | | Descriptor: | 2-fluoroadenine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3H3K
 
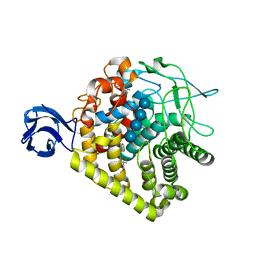 | |
3HSY
 
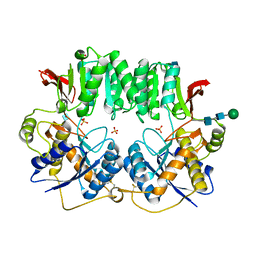 | | High resolution structure of a dimeric GluR2 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, SULFATE ION, ... | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
3IPD
 
 | | Helical extension of the neuronal SNARE complex into the membrane, spacegroup I 21 21 21 | | Descriptor: | Synaptosomal-associated protein 25, Syntaxin-1A, Vesicle-associated membrane protein 2 | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3GX2
 
 | |
3HAU
 
 | |
3GX6
 
 | |
3GZK
 
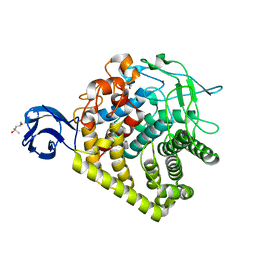 | | Structure of A. Acidocaldarius Cellulase CelA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cellulase, ... | | Authors: | Morera, S, Eckert, K, Vigouroux, A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of A. acidocaldarius endoglucanase Cel9A in complex with cello-oligosaccharides: strong -1 and -2 subsites mimic cellobiohydrolase activity
J.Mol.Biol., 394, 2009
|
|
3IWW
 
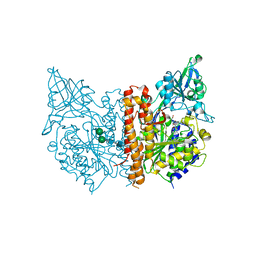 | |
3IYZ
 
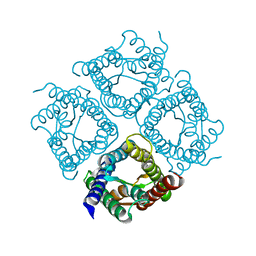 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
3IY8
 
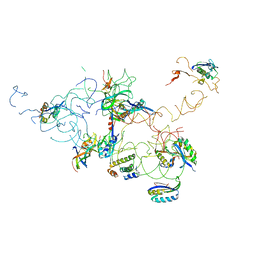 | | Leishmania tarentolae Mitonchondrial Ribosome small subunit | | Descriptor: | 30S ribosomal protein S11, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HDK
 
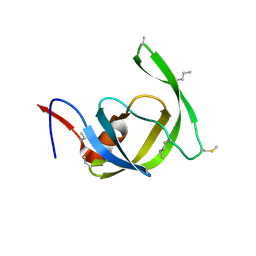 | | Crystal structure of chemically synthesized [Aib51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [Aib51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-28 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HD7
 
 | | HELICAL EXTENSION OF THE NEURONAL SNARE COMPLEX INTO THE MEMBRANE, spacegroup C 1 2 1 | | Descriptor: | SULFATE ION, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3J99
 
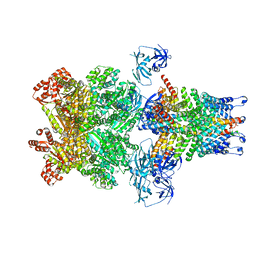 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
