4V90
 
 | | Thermus thermophilus Ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Chen, Y, Feng, S, Kumar, V, Ero, R, Gao, Y.G. | | Deposit date: | 2014-02-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of EF-G-ribosome complex in a pretranslocation state.
Nat. Struct. Mol. Biol., 20, 2013
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V5E
 
 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V4H
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4V8O
 
 | | Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3 | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-07-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Hybrid State of Ribosome in Complex with the Guanosine Triphosphatase Release Factor 3.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V8U
 
 | | Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.G, Feng, S, Chen, Y. | | Deposit date: | 2012-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex.
PLoS ONE, 8, 2013
|
|
4V5R
 
 | | The crystal structure of EF-Tu and Trp-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4UX1
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4V5A
 
 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4V5S
 
 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2AWO
 
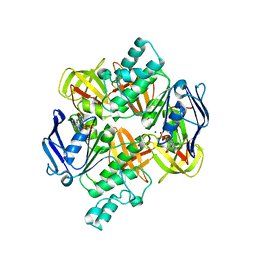 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ADP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AY5
 
 | |
2AZB
 
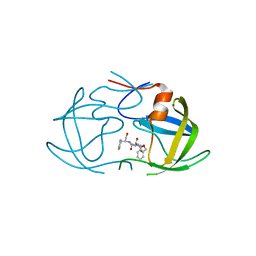 | | HIV-1 Protease NL4-3 3X mutant in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2AZJ
 
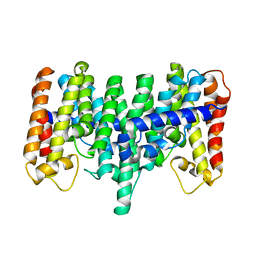 | | Crystal structure for the mutant D81C of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2B3P
 
 | | Crystal structure of a superfolder green fluorescent protein | | Descriptor: | ACETIC ACID, CADMIUM ION, green fluorescent protein | | Authors: | Pedelacq, J.D, Cabantous, S, Tran, T.H, Terwilliger, T.C, Waldo, G.S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and characterization of a superfolder green fluorescent protein.
Nat.Biotechnol., 24, 2006
|
|
2AK3
 
 | |
2B4S
 
 | | Crystal structure of a complex between PTP1B and the insulin receptor tyrosine kinase | | Descriptor: | Insulin receptor, SULFATE ION, Tyrosine-protein phosphatase, ... | | Authors: | Li, S, Depetris, R.S, Barford, D, Chernoff, J, Hubbard, S.R. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Complex between Protein Tyrosine Phosphatase 1B and the Insulin Receptor Tyrosine Kinase.
Structure, 13, 2005
|
|
2AIG
 
 | | ADAMALYSIN II WITH PEPTIDOMIMETIC INHIBITOR POL647 | | Descriptor: | ADAMALYSIN II, CALCIUM ION, N-(furan-2-ylcarbonyl)-L-leucyl-L-tryptophan, ... | | Authors: | Gomis-Rueth, F.X, Meyer, E.F, Kress, L.F, Politi, V. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of adamalysin II with peptidic inhibitors. Implications for the design of tumor necrosis factor alpha convertase inhibitors.
Protein Sci., 7, 1998
|
|
2AKP
 
 | | Hsp90 Delta24-N210 mutant | | Descriptor: | ATP-dependent molecular chaperone HSP82 | | Authors: | Richter, K, Moser, S, Hagn, F, Friedrich, R, Hainzl, O, Heller, M, Schlee, S, Kessler, H, Reinstein, J, Buchner, J. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Intrinsic inhibition of the Hsp90 ATPase activity.
J.Biol.Chem., 281, 2006
|
|
2AQT
 
 | | CU/ZN superoxide dismutase from neisseria meningitidis K91Q, K94Q double mutant | | Descriptor: | COPPER (I) ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | DiDonato, M, Kassmann, C.J, Bruns, C.K, Cabelli, D.E, Cao, Z, Tabatabai, L.B, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 2005-08-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CU/ZN superoxide dismutase from neisseria meningitidis K91Q, K94Q double mutant
To be Published
|
|
2ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Schuller, D.J, Gajhede, M. | | Deposit date: | 1997-08-19 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural interactions between horseradish peroxidase C and the substrate benzhydroxamic acid determined by X-ray crystallography.
Biochemistry, 37, 1998
|
|
2AUG
 
 | | Crystal structure of the Grb14 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 14 | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
2AX7
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AY2
 
 | |
