8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4K64
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8H2G
 
 | |
5ZVW
 
 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5ZS0
 
 | |
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | Descriptor: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1SBZ
 
 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
8H35
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.41 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3M7S
 
 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
5ZUZ
 
 | |
3DO3
 
 | | Human 1gG1 Fc fragment, 2.5 Angstrom structure | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Braden, B.C. | | Deposit date: | 2008-07-03 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human 1gG1 Fc fragment, 2.5 Angstrom structure.
To be Published
|
|
6A5N
 
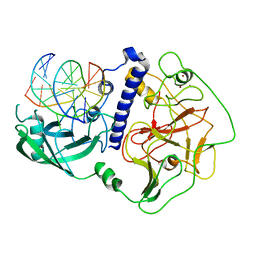 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZWN
 
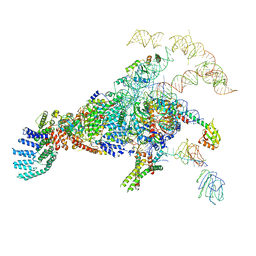 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.3 angstrom (Part II: U1 snRNP region) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Pre-mRNA-splicing ATP-dependent RNA helicase PRP28, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZZA
 
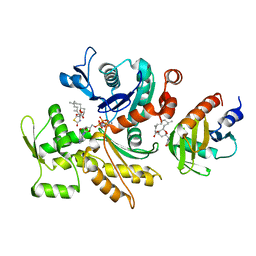 | | OdinProfilin/Rabbit Actin Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Genomes of Asgard archaea encode profilins that regulate actin.
Nature, 562, 2018
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6AH0
 
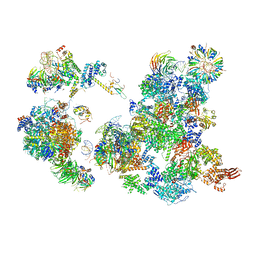 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
4KGG
 
 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
1SJV
 
 | | Three-Dimensional Structure of a Llama VHH Domain Swapping | | Descriptor: | r9 | | Authors: | Spinelli, S, Desmyter, A, Frenken, L, Verrips, T, Cambillau, C. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure.
Febs Lett., 564, 2004
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
3MA7
 
 | | Crystal structure of Cardiolipin bound to mouse CD1D | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2010-03-23 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Cardiolipin Binds to CD1d and Stimulates CD1d-Restricted {gamma}{delta} T Cells in the Normal Murine Repertoire.
J.Immunol., 186, 2011
|
|
