6NJ9
 
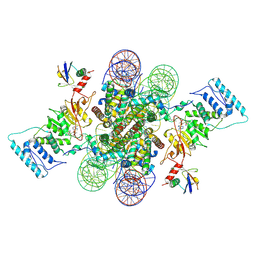 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 2-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-02 | | Release date: | 2019-02-20 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
1Y3D
 
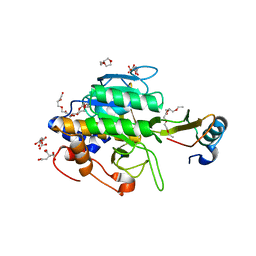 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 R67A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
5CTQ
 
 | |
6NN6
 
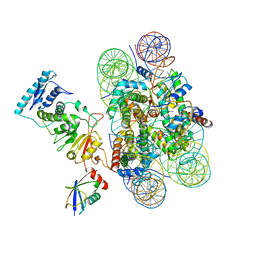 | | Structure of Dot1L-H2BK120ub nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Anderson, C.J, Baird, M.R, Hsu, A, Barbour, E.H, Koyama, Y, Borgnia, M.J, McGinty, R.K. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Cell Rep, 26, 2019
|
|
1NOV
 
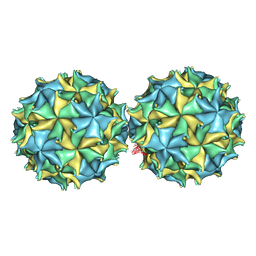 | | NODAMURA VIRUS | | Descriptor: | NODAMURA VIRUS COAT PROTEINS | | Authors: | Natarajan, P, Johnson, J.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4AQY
 
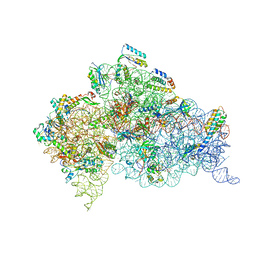 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7LA9
 
 | |
8E00
 
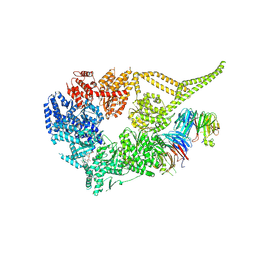 | | Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6Q88
 
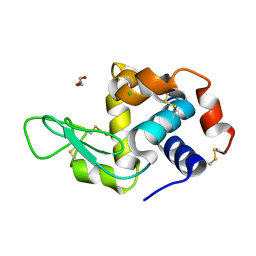 | | RT structure of HEWL at 5 kGy | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C | | Authors: | de la Mora, E, Coquelle, N, Bury, C.S, Rosenthal, M, Garman, E.F, Burghammer, M, Colletier, J.P, Weik, M. | | Deposit date: | 2018-12-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74007058 Å) | | Cite: | Radiation damage and dose limits in serial synchrotron crystallography at cryo- and room temperatures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1G1R
 
 | | Crystal structure of P-selectin lectin/EGF domains complexed with SLeX | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Somers, W.S, Camphausen, R.T. | | Deposit date: | 2000-10-13 | | Release date: | 2001-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1.
Cell(Cambridge,Mass.), 103, 2000
|
|
6O1K
 
 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:2:2 complex (core complex) | | Descriptor: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | Authors: | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | Deposit date: | 2019-02-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Architectural principles for Hfq/Crc-mediated regulation of gene expression
Elife, 8, 2019
|
|
6GMC
 
 | | 1.2 A resolution structure of human hydroxyacid oxidase 1 bound with FMN and 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, ... | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Smee, C, Arrowsmith, C.H, Edwards, E, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate
To Be Published
|
|
7LZ3
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5DEN
 
 | |
7Q3B
 
 | | Crystal structure of human STING in complex with 3'3'-c-(2'F,2'dA-isonucA)MP | | Descriptor: | 3'3'-c-(2'F,2'dA-isonucA)MP, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55733919 Å) | | Cite: | Discovery of isonucleotidic CDNs as potent STING agonists with immunomodulatory potential.
Structure, 30, 2022
|
|
6PST
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5b) with TraR and mutant rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
5EEU
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 1.31 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7Q0T
 
 | | Lysozyme soaked with V(IV)OSO4 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Fernandes, A.C.P, Correia, I, Sciortino, G, Garribba, E, Santos-Silva, T, Pessoa, J.C. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Chemistry, 28, 2022
|
|
1GG8
 
 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | Descriptor: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | Deposit date: | 2000-07-30 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
8EFS
 
 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
6PSV
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TpreRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6NTW
 
 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6CSO
 
 | | Crystal structure of the designed light-gated anion channel iC++ at pH6.5 | | Descriptor: | OLEIC ACID, RETINAL, iC++ | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
7TPQ
 
 | |
7O0T
 
 | | Crystal structure of Chloroflexus aggregans ene-reductase CaOYE holoenzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONATE ION, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Robescu, M.S, Niero, M, Loprete, G, Bergantino, E, Cendron, L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A New Thermophilic Ene-Reductase from the Filamentous Anoxygenic Phototrophic Bacterium Chloroflexus aggregans .
Microorganisms, 9, 2021
|
|
