5IC5
 
 | | Bacteriophytochrome response regulator RtBRR | | Descriptor: | CACODYLATE ION, Candidate response regulator, CheY, ... | | Authors: | Baker, A.W, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arm-in-Arm Response Regulator Dimers Promote Intermolecular Signal Transduction.
J.Bacteriol., 198, 2016
|
|
6K8E
 
 | | Global regulatory element SarX | | Descriptor: | HTH-type transcriptional regulator SarX | | Authors: | Wang, Q. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of SarX from Staphylococcus aureus
To Be Published
|
|
1GSN
 
 | | HUMAN GLUTATHIONE REDUCTASE MODIFIED BY DINITROSOGLUTATHIONE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLUTATHIONE REDUCTASE, ... | | Authors: | Becker, K, Savvides, S.N, Keese, M, Schirmer, R.H, Karplus, P.A. | | Deposit date: | 1998-02-21 | | Release date: | 1998-05-27 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat.Struct.Biol., 5, 1998
|
|
7CKK
 
 | | Structural complex of FTO bound with Dac51 | | Descriptor: | 2-{[2,6-dichloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino}-N-hydroxybenzamide, Alpha-ketoglutarate-dependent dioxygenase FTO, N-OXALYLGLYCINE | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance.
Cell Metab., 33, 2021
|
|
7E1B
 
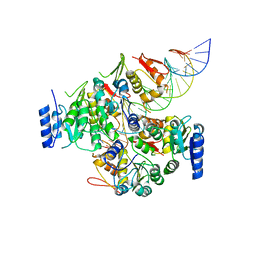 | | Crystal structure of VbrR-DNA complex | | Descriptor: | DNA (26-MER), DNA-binding response regulator | | Authors: | Hong, S, Zhang, X, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (4.587 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
8YPJ
 
 | |
2FEJ
 
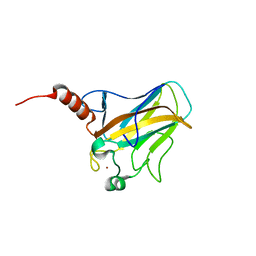 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GNV
 
 | |
5BRJ
 
 | |
8I74
 
 | |
8I75
 
 | |
8I76
 
 | |
5DYM
 
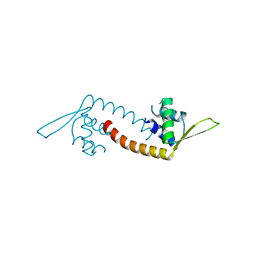 | | Crystal structure of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291 - CdPadR_0991 to 1.89 Angstrom resolution | | Descriptor: | PadR-family transcriptional regulator | | Authors: | Isom, C.E, Karr, E.A, Menon, S.K, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Crystal structure and DNA binding activity of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291.
Bmc Microbiol., 16, 2016
|
|
5TN2
 
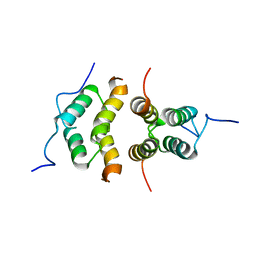 | | Solution Structure of the C-terminal multimerization domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
7SRV
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase (inactive form), spacegroup P22121 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
3EPS
 
 | |
5VAR
 
 | |
3DTU
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
7CFK
 
 | |
2RD8
 
 | |
7T3V
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase, spacegroup P22121 after crystals soaked with Zn2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
3IAU
 
 | | The structure of the processed form of threonine deaminase isoform 2 from Solanum lycopersicum | | Descriptor: | ACETATE ION, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Adaptive evolution of threonine deaminase in plant defense against insect herbivores.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2CJ5
 
 | |
2CJ8
 
 | |
2CJ4
 
 | |
