5TJA
 
 | | I-II linker of TRPML1 channel at pH 6 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7W7G
 
 | |
2VAY
 
 | | Calmodulin complexed with CaV1.1 IQ peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CHLORIDE ION, ... | | Authors: | Halling, D.B, Black, D.J, Pedersen, S.E, Hamilton, S.L. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Determinants in Cav1 Channels that Regulate the Ca2+ Sensitivity of Bound Calmodulin.
J.Biol.Chem., 284, 2009
|
|
3EFF
 
 | | The Crystal Structure of Full-Length KcsA in its Closed Conformation | | Descriptor: | FAB, Voltage-gated potassium channel | | Authors: | Uysal, S, Vasquez, V, Tereshko, T, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OVN
 
 | | Voltage-gated Sodium Channel 1.5 (Nav1.5) C-terminal domain in complex with Calmodulin poised for activation | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Boto, A, Jakoncic, J, Tomaselli, G.F, Amzel, L.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Nat Commun, 5, 2014
|
|
8QA1
 
 | |
8QA0
 
 | |
8D3Y
 
 | | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3X
 
 | | Human alpha3 Na+/K+-ATPase in its K+-occluded state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3U
 
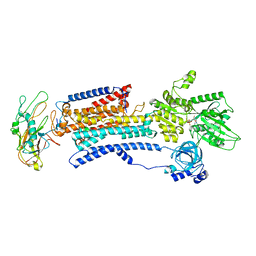 | | Human alpha3 Na+/K+-ATPase in its Na+-occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3W
 
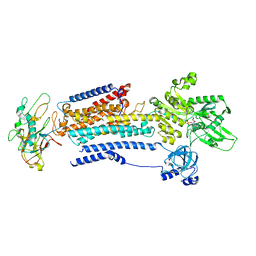 | |
8D3V
 
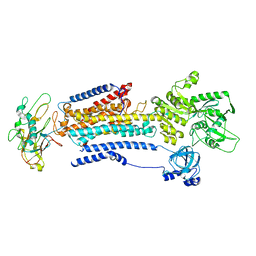 | | Human alpha3 Na+/K+-ATPase in its cytoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
6MUD
 
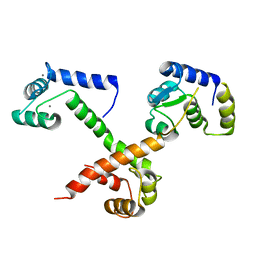 | |
6MUE
 
 | |
6DAH
 
 | | 2.5 Angstrom crystal structure of the N97S CaM mutant | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2KJ1
 
 | |
6BGJ
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
3J41
 
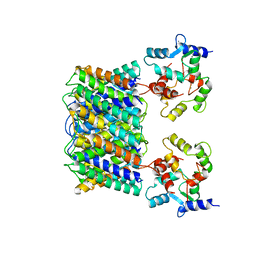 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8Q9Z
 
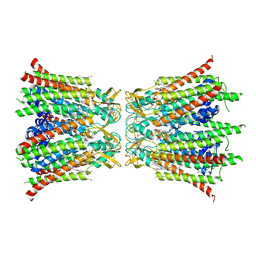 | |
6YHY
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conk-S1 | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6CSM
 
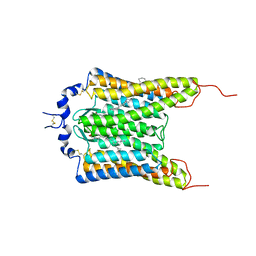 | | Crystal structure of the natural light-gated anion channel GtACR1 | | Descriptor: | GtACR1, OLEIC ACID, RETINAL | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
1WT7
 
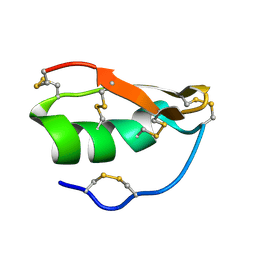 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
4DX1
 
 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
2N1N
 
 | | Solution structure of VSTx1 | | Descriptor: | Kappa-theraphotoxin-Gr3a | | Authors: | Lau, H.Y, King, G.F, Mobli, M. | | Deposit date: | 2015-04-12 | | Release date: | 2016-03-02 | | Last modified: | 2016-10-12 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the interaction between gating modifier spider toxins and the voltage sensor of voltage-gated ion channels.
Sci Rep, 6
|
|
