6T14
 
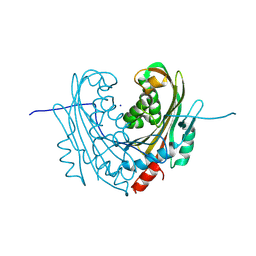 | | Native structure of mosquitocidal Cyt1A protoxin obtained by Serial Femtosecond Crystallography on in vivo grown crystals at pH 7 | | Descriptor: | SODIUM ION, Type-1Aa cytolytic delta-endotoxin | | Authors: | Tetreau, G, Banneville, A.S, Andreeva, E, Brewster, A.S, Hunter, M.S, Sierra, R.G, Young, I.D, Boutet, S, Coquelle, N, Cascio, D, Sawaya, M.R, Sauter, N.K, Colletier, J.P. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade.
Nat Commun, 11, 2020
|
|
3MA5
 
 | | Crystal structure of the tetratricopeptide repeat domain protein Q2S6C5_SALRD from Salinibacter ruber. Northeast Structural Genomics Consortium Target SrR115c. | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the tetratricopeptide repeat domain protein Q2S6C5_SALRD from Salinibacter ruber.
To be Published
|
|
3RBA
 
 | | Phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis complexed with DPCoA | | Descriptor: | DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2011-03-29 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Three-Dimensional Structure of Phosphopantetheine Adenylyltransferase
from Mycobacterium Tuberculosis in the Apo Form and in Complexes
with Coenzyme A and Dephosphocoenzyme A
CRYSTALLOGRAPHY REPORTS, 57, 2012
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
4EYT
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | SULFATE ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
7A4G
 
 | |
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
4K2X
 
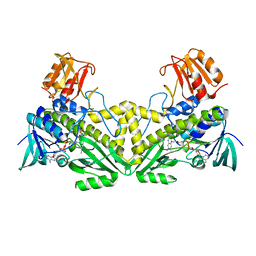 | |
4J8S
 
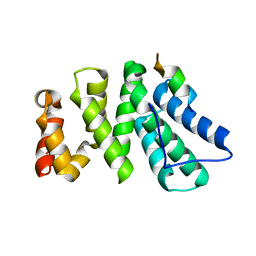 | | Crystal structure of human CNOT1 MIF4G domain in complex with a TTP peptide | | Descriptor: | CCR4-NOT transcription complex subunit 1, Tristetraprolin | | Authors: | Frank, F, Fabian, M.R, Rouya, C, Siddiqui, N, Lai, W.S, Karetnikov, A, Blackshear, P.J, Sonenberg, N, Nagar, B. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3CTB
 
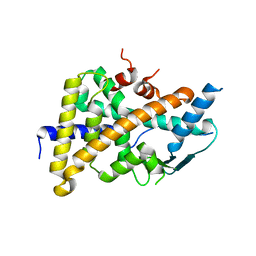 | | Tethered PXR-LBD/SRC-1p apoprotein | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2008-04-11 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
4D5C
 
 | |
7A4J
 
 | | Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-4 | | Descriptor: | Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase,6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Tetter, S, Hilvert, D. | | Deposit date: | 2020-08-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.
Science, 372, 2021
|
|
7A4H
 
 | |
7A4F
 
 | |
7A4I
 
 | | Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-3 | | Descriptor: | Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase,6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Tetter, S, Hilvert, D. | | Deposit date: | 2020-08-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.04 Å) | | Cite: | Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.
Science, 372, 2021
|
|
7URW
 
 | |
2KCV
 
 | | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c
To be Published
|
|
3MVP
 
 | | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A | | Descriptor: | TetR/AcrR transcriptional regulator | | Authors: | Stein, A.J, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A
To be Published
|
|
2KCL
 
 | | Solution NMR structure of tetratricopeptide repeat domain protein SrU_0103 from Salinibacter ruber, Northeast Structural Genomics Consortium (NESG) Target SrR115C | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of tetratricopeptide repeat domain protein SrU_0103 from Salinibacter ruber, Northeast Structural Genomics Consortium (NESG) Target SrR115C
To be Published
|
|
4LYM
 
 | |
4HKU
 
 | |
2M21
 
 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
1IES
 
 | | TETRAGONAL CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6OOL
 
 | | Structural elucidation of the Ectodomain of mouse UNC5H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Netrin receptor UNC5B, ... | | Authors: | Stetefeld, J, McDougall, M.D, Loewen, P.C, Moya, A, Meier, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural elucidation of the Ectodomain of mouse UNC5H2
To be published
|
|
6HX8
 
 | | Tubulin-STX3451 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dohle, W, Prota, A.E, Menchon, G, Hamel, E, Steinmetz, M.O, Potter, B.V.L. | | Deposit date: | 2018-10-16 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Tetrahydroisoquinoline Sulfamates as Potent Microtubule Disruptors: Synthesis, Antiproliferative and Antitubulin Activity of Dichlorobenzyl-Based Derivatives, and a Tubulin Cocrystal Structure.
ACS Omega, 4, 2019
|
|
