8Q7V
 
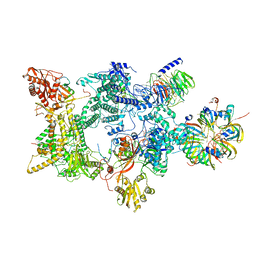 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 1) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8KG2
 
 | |
8Q7X
 
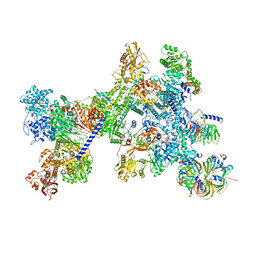 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 4) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7W
 
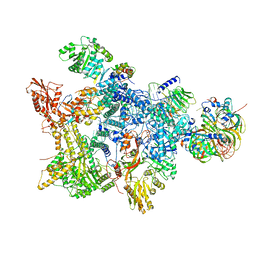 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 3) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TUI
 
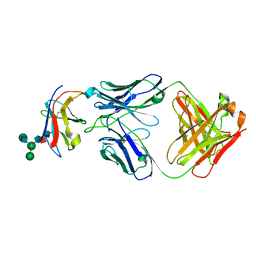 | | Crystal structure of Fab-Lirilumab bound to KIR2DL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell immunoglobulin-like receptor 2DL3, Lirilumab Fab Heavy Chain, ... | | Authors: | Lorig-Roach, N, DuBois, R.M. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activity and specificity of the immune checkpoint inhibitor lirilumab.
Sci Rep, 14, 2024
|
|
8TUC
 
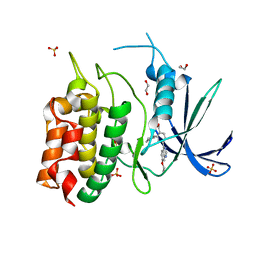 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
8Q7N
 
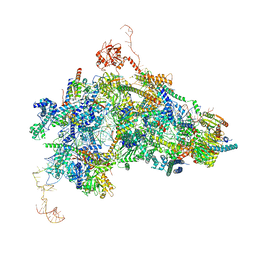 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
8KFP
 
 | |
8KFX
 
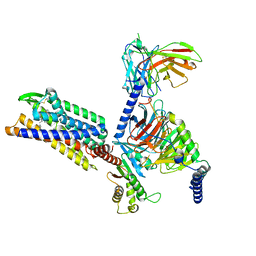 | | Gi bound CCR8 complex with nonpeptide agonist LMD-009 | | Descriptor: | 8-[[3-(2-methoxyphenoxy)phenyl]methyl]-1-(2-phenylethyl)-1,3,8-triazaspiro[4.5]decan-4-one, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFZ
 
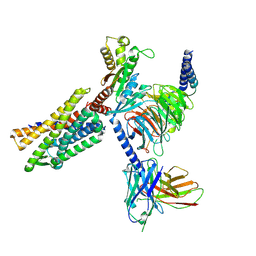 | | Gi bound CCR8 in ligand free state | | Descriptor: | C-C chemokine receptor type 8,LgBiT fusion protein,Recombinant Human Rhinovirus, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFY
 
 | | Gi bound CCR8 complex with nonpeptide agonist ZK 756326 | | Descriptor: | 2-[2-[4-[(3-phenoxyphenyl)methyl]piperazin-1-yl]ethoxy]ethanol, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8TUN
 
 | | S. thermodepolymerans KpsM-KpsE in Glycolipid 1 state with rigid body fitted KpsT | | Descriptor: | (2R,5S,8S)-2,5-dihydroxy-5,10-dioxo-8-[(undecanoyloxy)methyl]-4,6,9-trioxa-5lambda~5~-phosphahenicosan-1-yl 3-deoxy-alpha-L-altro-oct-2-ulopyranosidonic acid, ABC transporter ATP-binding protein, Capsular biosynthesis protein, ... | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 628, 2024
|
|
8Q7Q
 
 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 2) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q77
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BISUBSTRATE INHIBITOR ARC-780 | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[12-[[4-[[5-(4-carboxyphenyl)-1,3-thiazol-2-yl]amino]-4-oxidanylidene-butanoyl]-(2-hydroxy-2-oxoethyl)amino]dodecanoylamino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]butanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.255 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
8Q6X
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8Q6Y
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae in complex with cYY and Hypoxanthine | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, 1,2-ETHANEDIOL, Cytochrome P450, ... | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8TU6
 
 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8KFH
 
 | |
8KFI
 
 | |
8KFJ
 
 | |
8KF5
 
 | |
8KF4
 
 | |
8KF3
 
 | |
8KF6
 
 | |
8KF1
 
 | |
