7LCC
 
 | | Helitron transposase bound to LTS | | Descriptor: | Helraiser K1068Q, LTS, ZINC ION | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2021-01-10 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.
Mol.Cell, 81, 2021
|
|
3FCF
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-[(1E,7E)-8-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,6-dioxa-2,7-diazaocta-1,7-dien-1-yl]benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
8GJA
 
 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJ8
 
 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
1G9Y
 
 | | HOMING ENDONUCLEASE I-CREI / DNA SUBSTRATE COMPLEX WITH CALCIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
8KA4
 
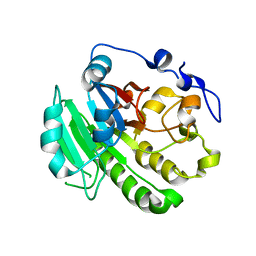 | |
5T3L
 
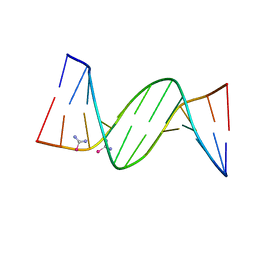 | | B-DNA (CGCGAATTCGCG)2 soaked with selenourea for 1 min | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), selenourea | | Authors: | Luo, Z, Dauter, Z. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Selenourea: a convenient phasing vehicle for macromolecular X-ray crystal structures.
Sci Rep, 6, 2016
|
|
3FCL
 
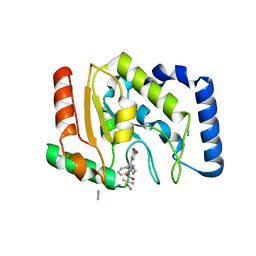 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{[(4-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}butyl)amino]methyl}benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
1G9Z
 
 | | LAGLIDADG HOMING ENDONUCLEASE I-CREI / DNA PRODUCT COMPLEX WITH MAGNESIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
3MIP
 
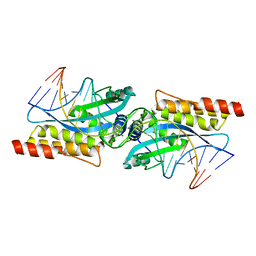 | | I-MsoI re-designed for altered DNA cleavage specificity (-8GCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*CP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-11 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
1QSL
 
 | |
2HC7
 
 | | 2'-selenium-T A-DNA [G(TSe)GTACAC] | | Descriptor: | 5'-D(*GP*(2ST)P*GP*TP*AP*CP*AP*C)-3' | | Authors: | Jiang, J, Sheng, J, Huang, Z. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of 2'-Selenium modified T A-DNA G(TSe)GTACAC
TO BE PUBLISHED
|
|
1ZZF
 
 | |
2PI2
 
 | | Full-length Replication protein A subunits RPA14 and RPA32 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation.
J.Mol.Biol., 374, 2007
|
|
2H7F
 
 | | Structure of variola topoisomerase covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
3OIY
 
 | | Helicase domain of reverse gyrase from Thermotoga maritima | | Descriptor: | CHLORIDE ION, PYROPHOSPHATE 2-, reverse gyrase helicase domain | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The latch modulates nucleotide and DNA binding to the helicase-like domain of Thermotoga maritima reverse gyrase and is required for positive DNA supercoiling.
Nucleic Acids Res., 39, 2011
|
|
1W3S
 
 | | The crystal structure of RecO from Deinococcus radiodurans. | | Descriptor: | HYPOTHETICAL PROTEIN DR0819, ZINC ION | | Authors: | Leiros, I, Timmins, J, Hall, D.R, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-18 | | Release date: | 2005-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and DNA-Binding Analysis of Reco from Deinococcus Radiodurans
Embo J., 24, 2005
|
|
1IYJ
 
 | | STRUCTURE OF A BRCA2-DSS1 COMPLEX | | Descriptor: | Deleted in split hand/split foot protein 1, breast cancer susceptibility | | Authors: | Pavletich, N.P, Jeffrey, P.D, Yang, H.J. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
8TDW
 
 | | ssRNA bound SAMHD1 T open | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
8TDV
 
 | | ssRNA bound SAMHD1 T closed | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
2Z6K
 
 | | Crystal structure of full-length human RPA14/32 heterodimer | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Habel, J.E, Kabaleeswaran, V, Borgstahl, G.E. | | Deposit date: | 2007-08-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation
J.Mol.Biol., 374, 2007
|
|
6OZE
 
 | |
7KRW
 
 | |
7KO2
 
 | |
7KRT
 
 | | Restraining state of a truncated Hsp70 DnaK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, MAGNESIUM ION | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Conformational equilibria in allosteric control of Hsp70 chaperones.
Mol.Cell, 81, 2021
|
|
