5EE7
 
 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
5YU2
 
 | | Structure of Ribonuclease YabJ | | Descriptor: | Translation initiation inhibitor homologue | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel chlorination-induced ribonuclease YabJ fromStaphylococcus aureus.
Biosci. Rep., 38, 2018
|
|
5US1
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
7CKO
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation | | Descriptor: | 7-[methyl-(phenylmethyl)amino]-2-oxidanylidene-chromene-3-carboxylic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6WYI
 
 | | Crystal structure of EchA19, enoyl-CoA hydratase from Mycobacterium tuberculosis | | Descriptor: | EchA19, enoyl-CoA hydratase, MAGNESIUM ION | | Authors: | Bonds, A.C, Garcia-Diaz, M, Sampson, N.S. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Post-translational Succinylation ofMycobacterium tuberculosisEnoyl-CoA Hydratase EchA19 Slows Catalytic Hydration of Cholesterol Catabolite 3-Oxo-chol-4,22-diene-24-oyl-CoA.
Acs Infect Dis., 6, 2020
|
|
6WRM
 
 | | The interaction of chlorido(1,5-cyclooctadiene)([4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1,3-dimethyl-1H-imidazol-3-ide])rhodium(I) with HEWL after 1 month | | Descriptor: | (4-{2-[(tert-butoxycarbonyl)amino]-3-methoxy-3-oxopropyl}-1,3-dimethyl-2,3-dihydro-1H-imidazol-2-yl)rhodium, CHLORIDE ION, Lysozyme, ... | | Authors: | Sullivan, M.P, John, M, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Combined Spectroscopic and Protein Crystallography Study Reveals Protein Interactions of Rh I (NHC) Complexes at the Molecular Level.
Inorg.Chem., 59, 2020
|
|
8FTN
 
 | | E. coli ArnA dehydrogenase domain mutant - N492A | | Descriptor: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | Authors: | Sousa, M.C, Mitchell, M.E. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
5ELN
 
 | |
4I50
 
 | | Crystal structure of tubulin-stathmin-TTL-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Prota, A.E, Bargsten, K, Zurwerra, D, Field, J.J, Diaz, J.F, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents.
Science, 339, 2013
|
|
5V6C
 
 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae | | Descriptor: | GLYCEROL, Hemolysin-related protein | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
7C60
 
 | | Crystal structure of Keap1 in complex with monoethyl fumarate (MEF) | | Descriptor: | (~{Z})-4-ethoxy-4-oxidanylidene-but-2-enoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P, Krishnappa, G. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
6WP9
 
 | | AvaR1 bound to Avenolide | | Descriptor: | (5S)-5-[(6R)-6-hydroxy-6-methyl-5-oxooctyl]furan-2(5H)-one, AvaR1 | | Authors: | Kapoor, I, Olivares, P.J, Nair, S.K. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Elife, 9, 2020
|
|
7UKW
 
 | | EGFR(T790M/V948R) in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Pham, C.D, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
6X18
 
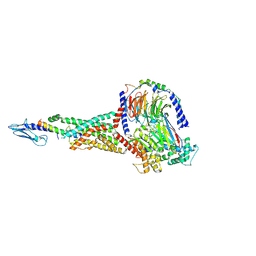 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
7CAB
 
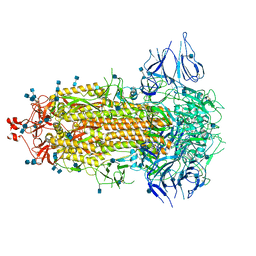 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
8G1Z
 
 | |
4I55
 
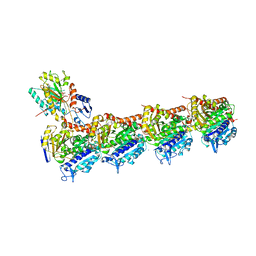 | | Crystal structure of tubulin-stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Prota, A.E, Bargsten, K, Zurwerra, D, Field, J.J, Diaz, J.F, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents.
Science, 339, 2013
|
|
8G20
 
 | |
5ESB
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
5ULG
 
 | |
7CM1
 
 | | Neuraminidase from the Wuhan Asiatic toad influenza virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Wang, J. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Lattice-translocation defects in specific crystals of the catalytic head domain of influenza neuraminidase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7UUY
 
 | | Structure of the sodium/iodide symporter (NIS) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Sodium/iodide cotransporter | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UUZ
 
 | | Structure of the sodium/iodide symporter (NIS) in complex with perrhenate and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, PERRHENATE, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
5ZH1
 
 | |
5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
