8D28
 
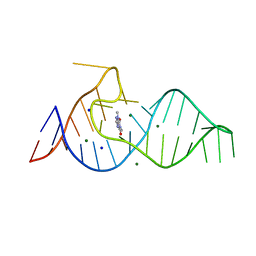 | |
8D2A
 
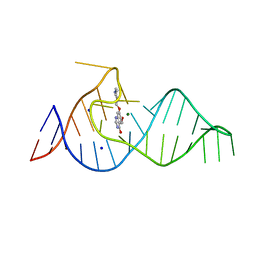 | |
8D5L
 
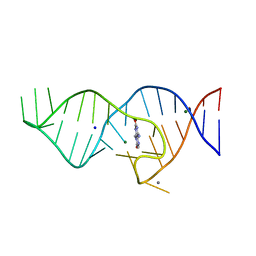 | | Crystal structure of theophylline aptamer in complex with TAL1 | | Descriptor: | 4-amino-8-methylpteridine-2,7(1H,8H)-dione, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Menichelli, E, Spraggon, G. | | Deposit date: | 2022-06-05 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of small molecules that target a tertiary-structured RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D29
 
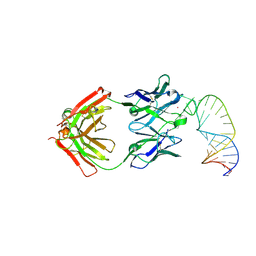 | |
2BII
 
 | | crystal structure of nitrate-reducing fragment of assimilatory nitrate reductase from Pichia angusta | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), GLYCEROL, NITRATE REDUCTASE [NADPH], ... | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
8D38
 
 | | Structure of a purine nucleoside phosphorylase from Geobacillus stearothermophilus | | Descriptor: | Purine nucleoside phosphorylase, SODIUM ION | | Authors: | Given, F, Johnston, J, Crittenden, D, Moran, F, Johns, A. | | Deposit date: | 2022-05-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The structure of His-tagged Geobacillus stearothermophilus purine nucleoside phosphorylase reveals a 'spanner in the works'.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5JDU
 
 | | Crystal structure for human thrombin mutant D189A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loop Electrostatics Asymmetry Modulates the Preexisting Conformational Equilibrium in Thrombin.
Biochemistry, 55, 2016
|
|
2DHO
 
 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
8D4O
 
 | |
8DJF
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound tetrahedral intermediate | | Descriptor: | (5S)-5-methyloxolane-2,2-diol, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-06-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DJZ
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound product | | Descriptor: | (4R)-4-hydroxypentanoic acid, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DJR
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJX
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - Compound II | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
6VFR
 
 | | Crystal structure of human protocadherin 18 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
8DJU
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - bicyclic dehydroascorbic acid complex | | Descriptor: | (3aS,6S,6aR)-3,3,3a,6-tetrahydroxytetrahydrofuro[3,2-b]furan-2(3H)-one (non-preferred name), L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJS
 
 | |
8DJT
 
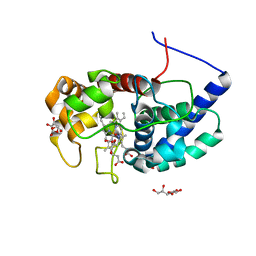 | |
8CX9
 
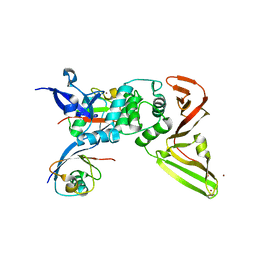 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
6V7Z
 
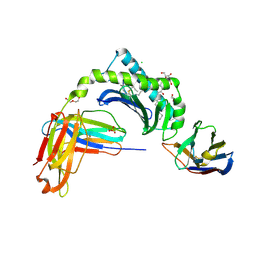 | | Human CD1d presenting alpha-Galactosylceramide in complex with VHH nanobody 1D22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A single-domain bispecific antibody targeting CD1d and the NKT T-cell receptor induces a potent antitumor response.
Nat Cancer, 2020
|
|
8DJW
 
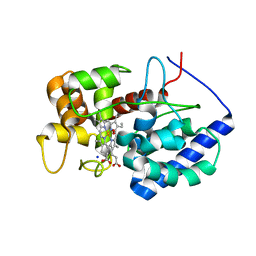 | |
5IUY
 
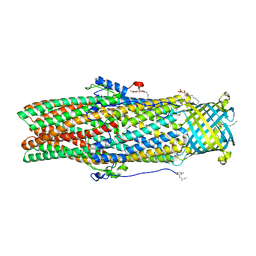 | | Structural insights of the outer-membrane channel OprN | | Descriptor: | CHLORIDE ION, FORMIC ACID, Multidrug efflux outer membrane protein OprN, ... | | Authors: | Ntsogo, Y, Garnier, C, Phan, G, Monlezun, L, Benas, P, Broutin, I. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
6V04
 
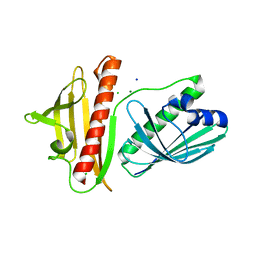 | | DynU16 crystal structure, a putative protein in the dynemicin biosynthetic locus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Alvarado, S.K, Miller, M.D, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of DynU16, a START/Bet v1-like protein involved in dynemicin biosynthesis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8D35
 
 | |
8DCM
 
 | |
5Y07
 
 | |
