8RSL
 
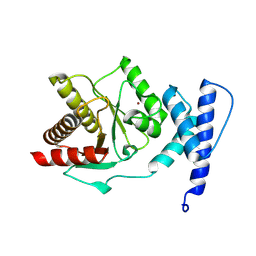 | | Crystal structure of Staphylococcus aureus macrodomain | | Descriptor: | Protein-ADP-ribose hydrolase, ZINC ION | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
8RSI
 
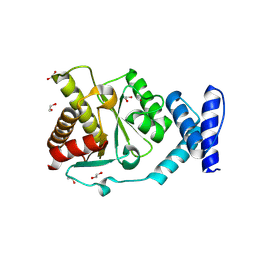 | | Crystal structure of Methanobrevibacter oralis macrodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
8RSN
 
 | |
5FSZ
 
 | |
5FUD
 
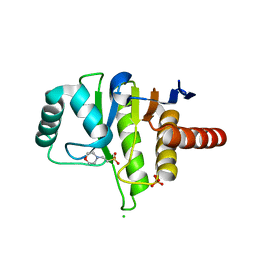 | | Oceanobacillus iheyensis macrodomain with MES bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, O-ACETYL-ADP-RIBOSE DEACETYLASE, ... | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis ofOceanobacillus iheyensismacrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5FSU
 
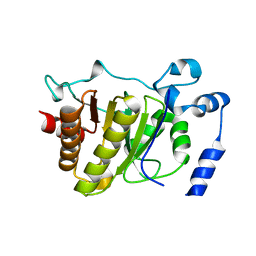 | |
5FSY
 
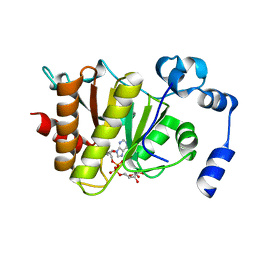 | | Crystal structure of Trypanosoma brucei macrodomain in complex with ADP-ribose | | Descriptor: | MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2016-01-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proximal Adp-Ribose Hydrolysis in Trypanosomatids is Catalyzed by a Macrodomain.
Sci.Rep., 6, 2016
|
|
5FSX
 
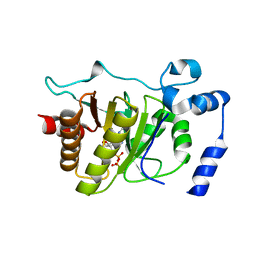 | |
5FSV
 
 | |
6Y73
 
 | | The crystal structure of human MACROD2 in space group P43 | | Descriptor: | ADP-ribose glycohydrolase MACROD2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KXB
 
 | | Crystal structure of SARS-CoV-2 Nsp3 Macrodomain complex with PARG329 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea, Non-structural protein 3, ... | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6Z72
 
 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6YWK
 
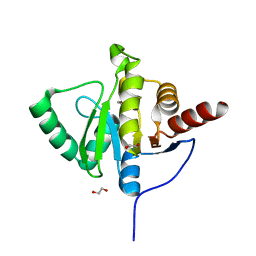 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7LG7
 
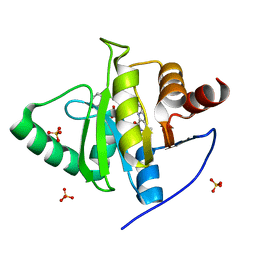 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | Descriptor: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6Z5T
 
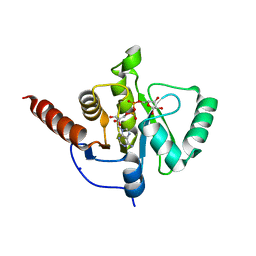 | | SARS-CoV-2 Macrodomain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab, SODIUM ION | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6Z6I
 
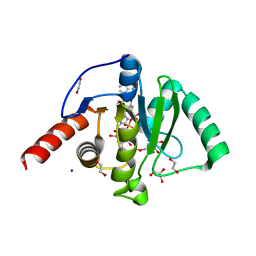 | | SARS-CoV-2 Macrodomain in complex with ADP-HPD | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7BF4
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7C33
 
 | | Macro domain of SARS-CoV-2 in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Structural, Biophysical, and Biochemical Elucidation of the SARS-CoV-2 Nonstructural Protein 3 Macro Domain.
Acs Infect Dis., 6, 2020
|
|
