8CRT
 
 | | Local refinement of Rh trimer, glycophorin B and Band3-III transmembrane region, class 1a of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, Band 3 anion transport protein, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OKP
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
8CTE
 
 | | Class 2 of erythrocyte ankyrin-1 complex (Composite map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, Ankyrin-1, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-14 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3U6I
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 58a | | Descriptor: | Hepatocyte growth factor receptor, N-{3-fluoro-4-[(7-methoxyquinolin-4-yl)oxy]phenyl}-1-[(2R)-2-hydroxypropyl]-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3GO5
 
 | |
1KIM
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
3HAW
 
 | |
4MLN
 
 | | Crystal of PhnZ bound to (R)-2-amino-1-hydroxyethylphosphonic acid | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6STB
 
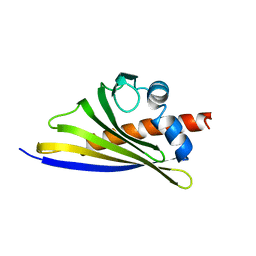 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
3U2T
 
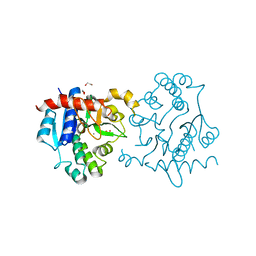 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U6H
 
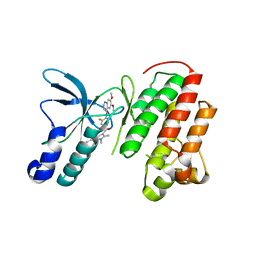 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3UBD
 
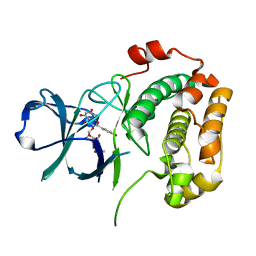 | | Structure of N-terminal domain of RSK2 kinase in complex with flavonoid glycoside SL0101 | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 3,4-di-O-acetyl-6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
5FA2
 
 | | Crystal structure of 426c.TM4deltaV1-3 p120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, THIOCYANATE ION, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
4Y6N
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and phosphoglyceric acid (PGA) - GpgS Mn2+ UDP-Glc PGA-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1KPM
 
 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
8CKC
 
 | | Vitamin D receptor complex with 25-amine derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R)-6-azanyl-6-methyl-heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
8CK5
 
 | | VDR LBD complex with 25-nitro derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-nitro-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
1KJ7
 
 | |
6DU3
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION, REST-pS861 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
1KJF
 
 | |
3TV1
 
 | | Crystal structure of RtcA.AMP product complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2011-09-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RNA 3'-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4MUH
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(2-(5-acetamido-1,3,4-thiadiazol-2-ylsulfonylcarbamoyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Pantothenate synthetase, ... | | Authors: | Silvestre, H.L, Blundell, T.L. | | Deposit date: | 2013-09-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of Inhibitors of Mycobacterium tuberculosis Pantothenate Synthetase Based on Group Efficiency Analysis.
Chemmedchem, 11, 2016
|
|
6ST9
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | Descriptor: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
7A3O
 
 | | Crystal structure of dengue 1 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | Core protein, GLYCEROL, SULFATE ION, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
1KLA
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
