3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KAA
 
 | | Structure of Tim-3 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Ballesteros, A, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells.
J.Immunol., 184, 2010
|
|
3JZ7
 
 | |
2WNG
 
 | | complete extracellular structure of human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Graham, S.C, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of Signal-Regulatory Protein Alpha: A Link to Antigen Receptor Evolution.
J.Biol.Chem., 284, 2009
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3HB3
 
 | | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Koepke, J, Angerer, H, Peng, G. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways
Biochim.Biophys.Acta, 1787, 2009
|
|
2WBW
 
 | | Ad37 fibre head in complex with CAR D1 and sialic acid | | Descriptor: | CALCIUM ION, COXSACKIEVIRUS AND ADENOVIRUS RECEPTOR, FIBER PROTEIN, ... | | Authors: | Seiradake, E, Henaff, D, Wodrich, H, Billet, O, Perreau, M, Hippert, C, Mennechet, F, Schoehn, G, Lortat-Jacob, H, Dreja, H, Ibanes, S, Kalatzis, V, Wang, J.P, Finberg, R.W, Cusack, S, Kremer, E.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Cell Adhesion Molecule "Car" and Sialic Acid on Human Erythrocytes Influence Adenovirus in Vivo Biodistribution.
Plos Pathog., 5, 2009
|
|
2W9L
 
 | | CANINE ADENOVIRUS TYPE 2 FIBRE HEAD IN COMPLEX WITH CAR DOMAIN D1 AND SIALIC ACID | | Descriptor: | COXSACKIEVIRUS AND ADENOVIRUS RECEPTOR, FIBRE PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Seiradake, E, Henaff, D, Wodrich, H, Billet, O, Perreau, M, Hippert, C, Mennechet, F, Schoehn, G, Lortat-Jacob, H, Dreja, H, Ibanes, S, Kalatzis, V, Wang, J.P, Finberg, R.W, Cusack, S, Kremer, E.J. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The cell adhesion molecule "CAR" and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution.
PLoS Pathog., 5, 2009
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
3EYO
 
 | | Crystal structure of anti-human cytomegalovirus antibody 8F9 | | Descriptor: | 8f9 Fab, AD-2 | | Authors: | Thomson, C.A, Bryson, S, McLean, G.R, Creagh, A.L, Pai, E.F, Schrader, J.W. | | Deposit date: | 2008-10-21 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus.
Embo J., 27, 2008
|
|
3EYU
 
 | |
3EYQ
 
 | | Crystal structure of MJ5 Fab, a germline antibody variant of anti-human cytomegalovirus antibody 8f9 | | Descriptor: | 8f9 Fab, M2J5 Fab | | Authors: | Thomson, C.A, Bryson, S, McLean, G.R, Creagh, A.L, Pai, E.F, Schrader, J.W. | | Deposit date: | 2008-10-21 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus.
Embo J., 27, 2008
|
|
3EYS
 
 | |
3EYF
 
 | | Crystal structure of anti-human cytomegalovirus antibody 8f9 plus gB peptide | | Descriptor: | 8f9 Fab, AD-2, GLYCEROL, ... | | Authors: | Thomson, C.A, Bryson, S, McLean, G.R, Creagh, A.L, Pai, E.F, Schrader, J.W. | | Deposit date: | 2008-10-20 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus.
Embo J., 27, 2008
|
|
3EOY
 
 | | Structure of Reovirus sigma1 in Complex with Its Receptor Junctional Adhesion Molecule-A | | Descriptor: | Junctional adhesion molecule A, Outer capsid protein sigma-1 | | Authors: | Kirchner, E, Guglielmi, K.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A
Plos Pathog., 4, 2008
|
|
3EPD
 
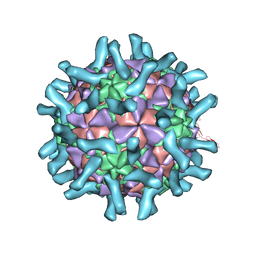 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPC
 
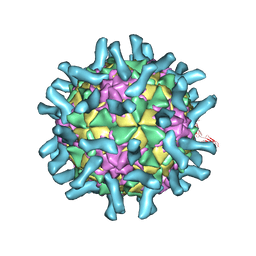 | | CryoEM structure of poliovirus receptor bound to poliovirus type 1 | | Descriptor: | MYRISTIC ACID, Poliovirus receptor, Protein VP1, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPF
 
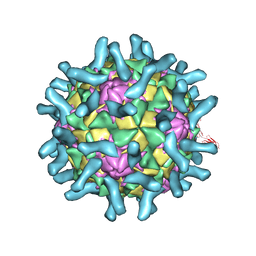 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EHB
 
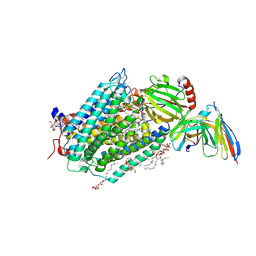 | | A D-Pathway Mutation Decouples the Paracoccus Denitrificans Cytochrome c Oxidase by Altering the side chain orientation of a distant, conserved Glutamate | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1-beta, ... | | Authors: | Koepke, J, Mueller, H, Peng, G. | | Deposit date: | 2008-09-12 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A d-pathway mutation decouples the paracoccusdenitrificans cytochrome C oxidase by altering the side-chain orientation of a distant conserved glutamate
J.Mol.Biol., 384, 2008
|
|
2W0L
 
 | |
2W0K
 
 | |
3E3Q
 
 | |
3E2H
 
 | |
3DV4
 
 | | Crystal structure of SAG506-01, tetragonal, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DVN
 
 | |
