3SHU
 
 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SJ7
 
 | |
3U6W
 
 | | Truncated M. tuberculosis LeuA (1-425) complexed with KIV | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, GLYCEROL, ... | | Authors: | Koon, N, Baker, E.N, Squire, C.J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Removal of the C-terminal regulatory domain of alpha-isopropylmalate synthase disrupts functional substrate binding.
Biochemistry, 51, 2012
|
|
2HL8
 
 | |
2HKP
 
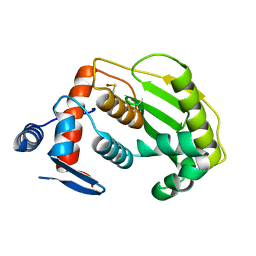 | |
3RB9
 
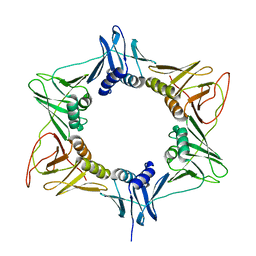 | |
3TD6
 
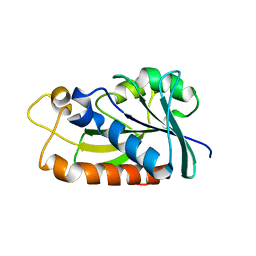 | | Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis from trigonal partially dehydrated crystal | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Ahmad, R, Varshney, U, Vijayan, M. | | Deposit date: | 2011-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of new crystal forms of Mycobacterium tuberculosis peptidyl-tRNA hydrolase and functionally important plasticity of the molecule
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3TFM
 
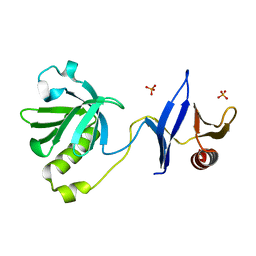 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
3UAM
 
 | |
3TK5
 
 | | Factor Xa in complex with D102-4380 | | Descriptor: | 4-{3-[(4-chlorophenyl)amino]-3-oxopropyl}-3-({[5-(propan-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]carbonyl}amino)benzoic acid, CALCIUM ION, Factor X heavy chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
3TZ6
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2GHI
 
 | | Crystal Structure of Plasmodium yoelii Multidrug Resistance Protein 2 | | Descriptor: | SULFATE ION, transport protein | | Authors: | Dong, A, Gao, M, Choe, J, Zhao, Y, Lew, J, Wasney, G, Alam, Z, Melone, M, Kozieradzki, I, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3QI1
 
 | |
3QKD
 
 | | Crystal structure of Bcl-xL in complex with a Quinazoline sulfonamide inhibitor | | Descriptor: | (R)-N-(7-(4-((4'-chlorobiphenyl-2-yl)methyl)piperazin-1-yl)quinazolin-4-yl)-4-(4-(dimethylamino)-1-(phenylthio)butan-2-ylamino)-3-nitrobenzenesulfonamide, Bcl-2-like protein 1, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Smith, B.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity.
J.Med.Chem., 54, 2011
|
|
3QVT
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus wild-type with the intermediate 5-keto 1-phospho glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
3Q6I
 
 | | Crystal structure of FabG4 and coenzyme binary complex | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-01-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of holoFabG4
To be Published
|
|
3QPY
 
 | | Crystal structure of a mutant (K57A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
3TKT
 
 | | Crystal structure of CYP108D1 from Novosphingobium aromaticivorans DSM12444 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2011-08-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of CYP108D1 from Novosphingobium aromaticivorans DSM12444: an aromatic hydrocarbon-binding P450 enzyme
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TOI
 
 | |
3TTU
 
 | | Structure of F413Y/H128N double variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3U4N
 
 | | A novel covalently linked insulin dimer | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Norrman, M, Vinther, T.N. | | Deposit date: | 2011-10-10 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel covalently linked insulin dimer engineered to investigate the function of insulin dimerization.
Plos One, 7, 2012
|
|
3UG8
 
 | | AKR1C3 complex with indomethacin at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, INDOMETHACIN, ... | | Authors: | Flanagan, J.U, Yosaatmadja, Y, Teague, R.M, Chai, M, Squire, C.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
4U6E
 
 | | HsMetAP in complex with (amino(phenyl)methyl)phosphonic acid | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 1, [(R)-amino(phenyl)methyl]phosphonic acid | | Authors: | Arya, T, Addlagatta, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the Molecular Basis of Inhibitor Selectivity between the Human and Streptococcal Type I Methionine Aminopeptidases
J.Med.Chem., 58, 2015
|
|
3LJF
 
 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
5WMF
 
 | |
