1H2H
 
 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
1H2I
 
 | | Human Rad52 protein, N-terminal domain | | Descriptor: | DNA REPAIR PROTEIN RAD52 HOMOLOG | | Authors: | Singleton, M.R, Wentzell, L.M, Liu, Y, West, S.C, Wigley, D.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Single-Strand Annealing Domain of Human Rad52 Protein
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H2J
 
 | | ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION | | Descriptor: | 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-BETA-D-CELLOBIOSIDE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Varrot, A, Davies, G.J. | | Deposit date: | 2002-08-09 | | Release date: | 2002-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1H2K
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2L
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FACTOR INHIBITING HIF1, FE (II) ION, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2M
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, N-OXALYLGLYCINE, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2N
 
 | | Factor Inhibiting HIF-1 alpha | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2O
 
 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2R
 
 | |
1H2S
 
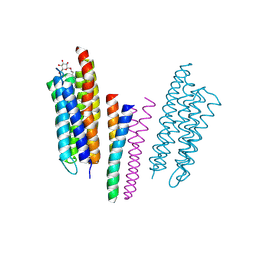 | | Molecular basis of transmenbrane signalling by sensory rhodopsin II-transducer complex | | Descriptor: | RETINAL, SENSORY RHODOPSIN II, SENSORY RHODOPSIN II TRANSDUCER, ... | | Authors: | Gordeliy, V.I, Labahn, J, Moukhametzianov, R, Efremov, R, Granzin, J, Schlesinger, R, Bueldt, G, Savopol, T, Scheidig, A, Klare, J.P, Engelhard, M. | | Deposit date: | 2002-08-15 | | Release date: | 2002-10-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular Basis of Transmembrane Signalling by Sensory Rhodopsin II-Transducer Complex
Nature, 419, 2002
|
|
1H2T
 
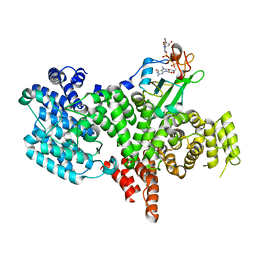 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2U
 
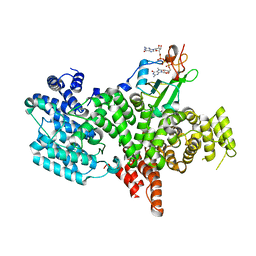 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2V
 
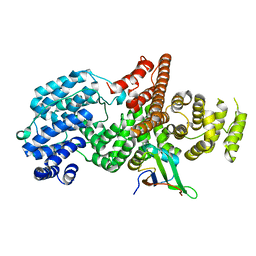 | | Structure of the human nuclear cap-binding-complex (CBC) | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 80 KDA NUCLEAR CAP BINDING PROTEIN | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2W
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1H2X
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, Y473F MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1H2Y
 
 | |
1H2Z
 
 | |
1H30
 
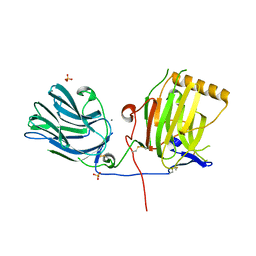 | | C-terminal LG domain pair of human Gas6 | | Descriptor: | CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN, SULFATE ION | | Authors: | Sasaki, T, Knyazev, P.G, Cheburkin, Y, Gohring, W, Tisi, D, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2002-08-21 | | Release date: | 2003-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Carboxy-Terminal Fragment of Growth-Arrest-Specific Protein Gas6: Receptor Tyrosine Kinase Activation by Laminin G-Like Domains
J.Biol.Chem., 277, 2002
|
|
1H31
 
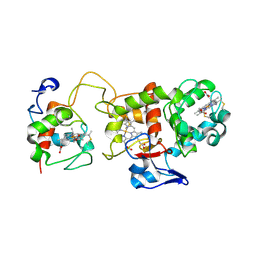 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H32
 
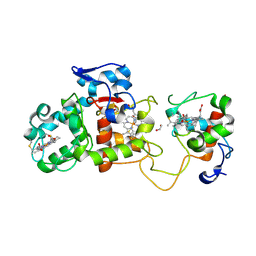 | | Reduced SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME C, DIHEME CYTOCHROME C, ... | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H33
 
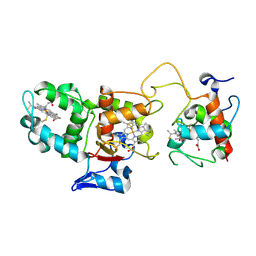 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H34
 
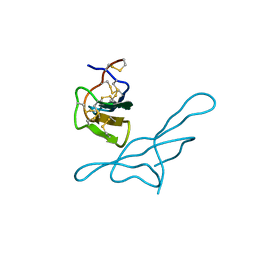 | | Crystal structure of lima bean trypsin inhibitor | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Debreczeni, J.E, Bunkoczi, G, Girmann, B, Sheldrick, G.M. | | Deposit date: | 2002-08-21 | | Release date: | 2003-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In-House Phase Determination of the Lima Bean Trypsin Inhibitor: A Low-Resolution Sulfur-Sad Case
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1H35
 
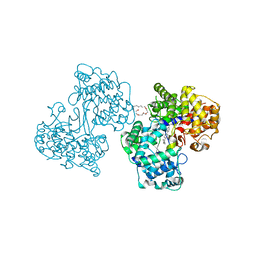 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4'-{[ALLYL(METHYL)AMINO]METHYL}-1,1'-BIPHENYL-4-YL)(4-BROMOPHENYL)METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
