4UWE
 
 | |
4UWA
 
 | |
3POU
 
 | |
3M9S
 
 | | Crystal structure of respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | Deposit date: | 2010-03-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The architecture of respiratory complex I
Nature, 465, 2010
|
|
3M9C
 
 | | Crystal structure of the membrane domain of respiratory complex I from Escherichia coli | | Descriptor: | NADH-quinone oxidoreductase subunit NuoL, NADH-quinone oxidoreductase subunit NuoM, NADH-quinone oxidoreductase subunit NuoN, ... | | Authors: | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | Deposit date: | 2010-03-22 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The architecture of respiratory complex I
Nature, 465, 2010
|
|
3POQ
 
 | |
3POX
 
 | |
3RKO
 
 | |
5T8A
 
 | | Recombinant cytotoxin-I from the venom of cobra N. oxiana | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
1IH9
 
 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
1H2S
 
 | | Molecular basis of transmenbrane signalling by sensory rhodopsin II-transducer complex | | Descriptor: | RETINAL, SENSORY RHODOPSIN II, SENSORY RHODOPSIN II TRANSDUCER, ... | | Authors: | Gordeliy, V.I, Labahn, J, Moukhametzianov, R, Efremov, R, Granzin, J, Schlesinger, R, Bueldt, G, Savopol, T, Scheidig, A, Klare, J.P, Engelhard, M. | | Deposit date: | 2002-08-15 | | Release date: | 2002-10-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular Basis of Transmembrane Signalling by Sensory Rhodopsin II-Transducer Complex
Nature, 419, 2002
|
|
1ZAD
 
 | | Structure of cytotoxin I (CTI) from Naja Oxiana in complex with DPC micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Konshina, A.G, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-06-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins
Biochem.J., 387, 2005
|
|
8OQY
 
 | |
8OQZ
 
 | |
1DXZ
 
 | | M2 TRANSMEMBRANE SEGMENT OF ALPHA-SUBUNIT OF NICOTINIC ACETYLCHOLINE RECEPTOR FROM TORPEDO CALIFORNICA, NMR, 20 STRUCTURES | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN | | Authors: | Pashkov, V.S, Maslennikov, I.V, Tchikin, L.D, Efremov, R.G, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 2000-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of the M2 Transmembrane Segment of the Nicotinic Acetylcholine Receptor Alpha-Subunit
FEBS Lett., 457, 1999
|
|
4O9Y
 
 | | Crystal Structure of TcdA1 | | Descriptor: | TcdA1 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
4O9X
 
 | | Crystal Structure of TcdB2-TccC3 | | Descriptor: | MERCURY (II) ION, TcdB2, TccC3 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
2PCO
 
 | | Spatial Structure and Membrane Permeabilization for Latarcin-1, a Spider Antimicrobial Peptide | | Descriptor: | Latarcin-1 | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity.
Biochemistry, 47, 2008
|
|
7QD5
 
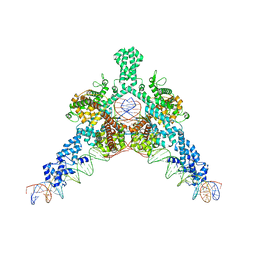 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA | | Descriptor: | IR48 DNA substrate, non transferred strand, IR48 transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD8
 
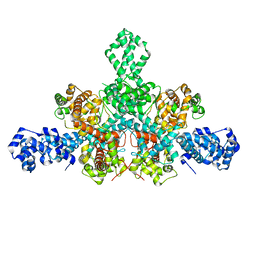 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state | | Descriptor: | Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD6
 
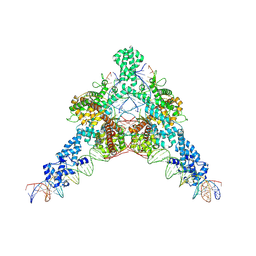 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product | | Descriptor: | IR71st non transferred strand, IR71st transferred strand, Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD4
 
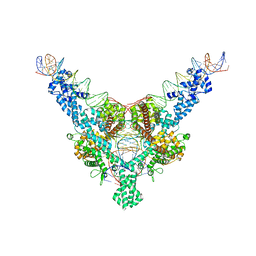 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA | | Descriptor: | IR100 DNA substrate, none transferred strand, transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
5NQ4
 
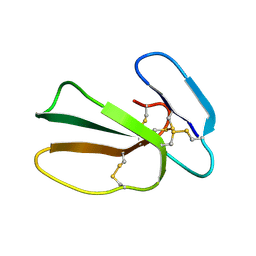 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
7Z0D
 
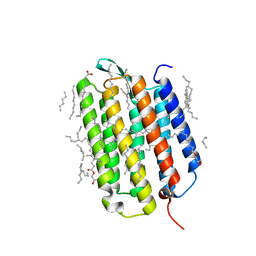 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z09
 
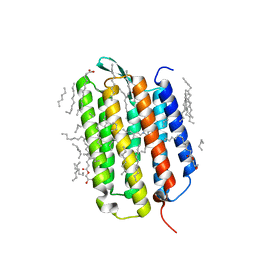 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
