8EW2
 
 | |
7TNY
 
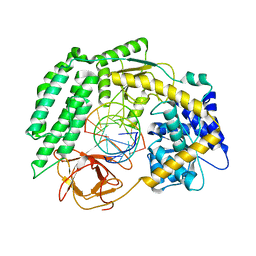 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNX
 
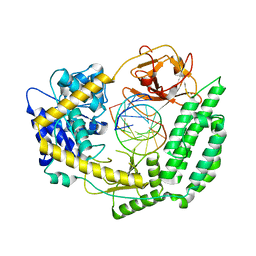 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO2
 
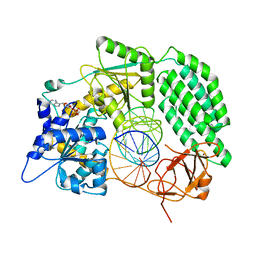 | |
7TO1
 
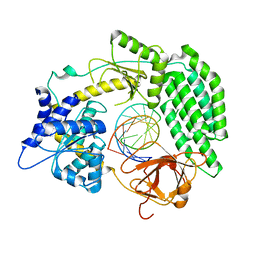 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+ATP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3SLR30 | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO0
 
 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VDC
 
 | |
8I8Y
 
 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7VDQ
 
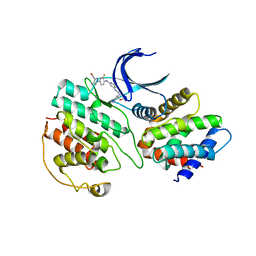 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 7 | | Descriptor: | 2-[[7-[[2-fluoranyl-4-[3-(hydroxymethyl)pyrazol-1-yl]phenyl]amino]-1,6-naphthyridin-2-yl]-(1-methylpiperidin-4-yl)amino]ethanoic acid, Cyclin-dependent kinase 5 activator 1, p25, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDS
 
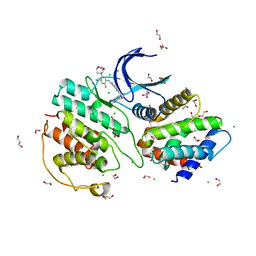 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 24 | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-4-[[2-[(1R)-1-(1-methylpiperidin-4-yl)-1-oxidanyl-ethyl]-1,6-naphthyridin-7-yl]amino]-2-morpholin-4-yl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDR
 
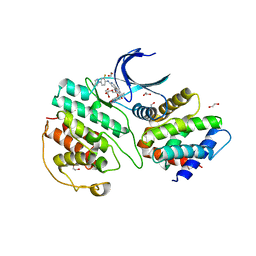 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 13 | | Descriptor: | (1R)-1-[7-[(2-fluoranyl-4-pyrazol-1-yl-phenyl)amino]-1,6-naphthyridin-2-yl]-1-(1-methylpiperidin-4-yl)ethanol, 1,2-ETHANEDIOL, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDP
 
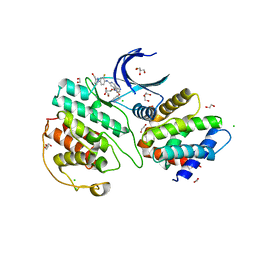 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
8OVS
 
 | |
8CDK
 
 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
4EVH
 
 | |
6W2S
 
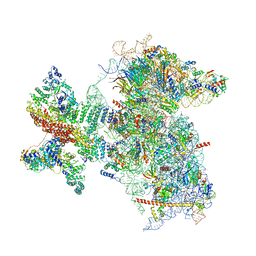 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
4EIV
 
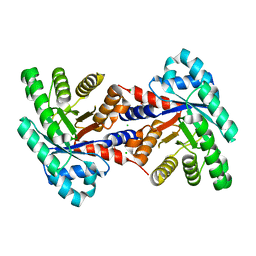 | | 1.37 Angstrom resolution crystal structure of apo-form of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Deoxyribose-phosphate aldolase | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
4UW1
 
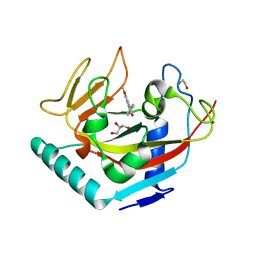 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(dimethylamino)methyl]phenyl}-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1-Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
4EVF
 
 | |
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4EL1
 
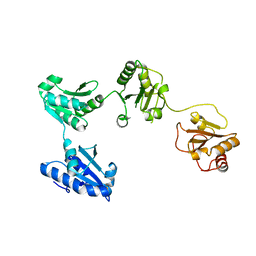 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
1SJ8
 
 | | Solution Structure of the R1R2 Domains of Talin | | Descriptor: | Talin 1 | | Authors: | Papagrigoriou, E, Gingras, A.R, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle
EMBO J., 23, 2004
|
|
