7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7M2X
 
 | | Open conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7MM1
 
 | | PTP1B in complex with TCS401 by Native S-SAD at Room Temperature | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-04-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7M2Z
 
 | | Monomeric single-particle reconstruction of the Yeast gamma-TuSC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component SPC97, Spindle pole body component SPC98, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
6QXP
 
 | | Protein peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat extensin-like protein 2, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2G4L
 
 | | Anomalous substructure of hydroxynitrile lyase | | Descriptor: | (S)-acetone-cyanohydrin lyase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5LA7
 
 | | Crystal structure of human proheparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5KWF
 
 | | Joint X-ray Neutron Structure of Cholesterol Oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Golden, E, Vrielink, A, Meilleur, F, Blakeley, M. | | Deposit date: | 2016-07-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.499 Å), X-RAY DIFFRACTION | | Cite: | An extended N-H bond, driven by a conserved second-order interaction, orients the flavin N5 orbital in cholesterol oxidase.
Sci Rep, 7, 2017
|
|
6L5Z
 
 | |
4V8C
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V87
 
 | | Crystal structure analysis of ribosomal decoding. | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-09-20 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
5DJ5
 
 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
8RU1
 
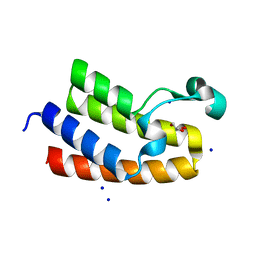 | | Chromatin remodeling regulator CECR2 with in crystallo disulfide bond | | Descriptor: | Chromatin remodeling regulator CECR2, GLYCEROL, SODIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
8RYS
 
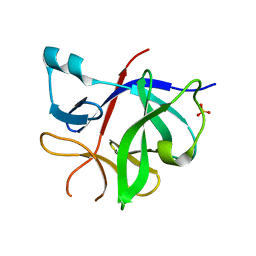 | | Human IL-1beta, unliganded | | Descriptor: | Interleukin-1 beta, SULFATE ION | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
2FPW
 
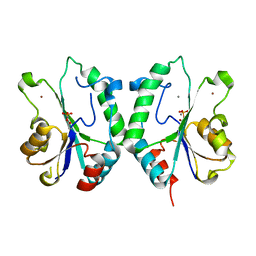 | | Crystal Structure of the N-terminal Domain of E.coli HisB- Phosphoaspartate intermediate. | | Descriptor: | CALCIUM ION, Histidine biosynthesis bifunctional protein hisB, ZINC ION | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
5CSH
 
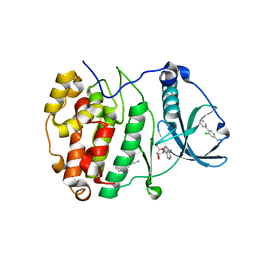 | | Crystal Structure of CK2alpha with Compound 4 bound | | Descriptor: | 1-(2-chlorobiphenyl-4-yl)methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8RC0
 
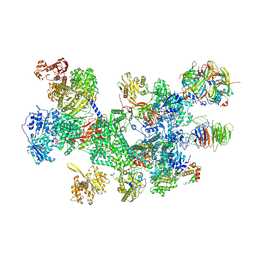 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
8RWU
 
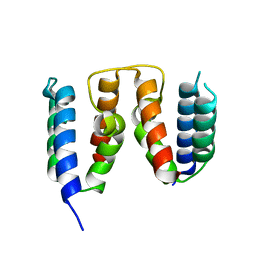 | |
2FQZ
 
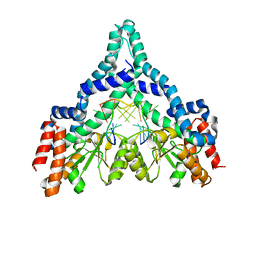 | | Metal-depleted Ecl18kI in complex with uncleaved DNA | | Descriptor: | DNA STRAND 1, DNA STRAND 2, R.Ecl18kI | | Authors: | Bochtler, M, Szczepanowski, R.H, Tamulaitis, G, Grazulis, S, Czapinska, H, Manakova, E, Siksnys, V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease
Embo J., 25, 2006
|
|
5D9D
 
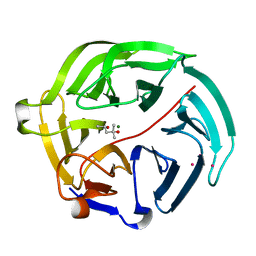 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
2G76
 
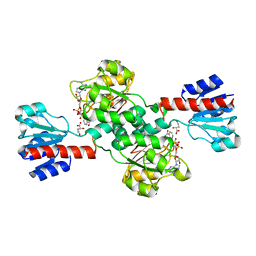 | | Crystal structure of human 3-phosphoglycerate dehydrogenase | | Descriptor: | D-3-phosphoglycerate dehydrogenase, D-MALATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Turnbull, A.P, Salah, E, Savitsky, P, Gileadi, O, von Delft, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase
To be Published
|
|
8RHN
 
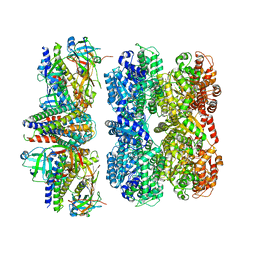 | | Structure of the 55LCC ATPase complex | | Descriptor: | ATPase family gene 2 protein homolog A, ATPase family gene 2 protein homolog B, Cyclin-dependent kinase 2-interacting protein, ... | | Authors: | Foglizzo, M, Degtjarik, O, Zeqiraj, E. | | Deposit date: | 2023-12-15 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
