7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3L9N
 
 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3LNH
 
 | |
7S5X
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S61
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5V
 
 | | Human KATP channel in open conformation, focused on SUR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Y
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NAS
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7S5Z
 
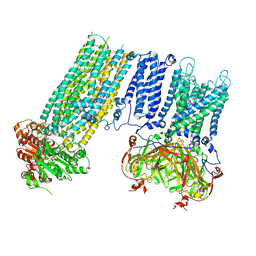 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N30
 
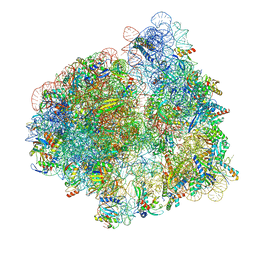 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7S60
 
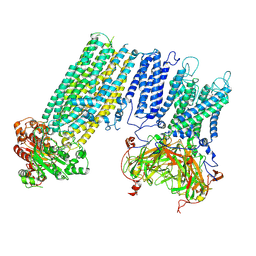 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5GW1
 
 | |
3L9L
 
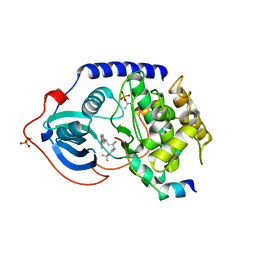 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1RY4
 
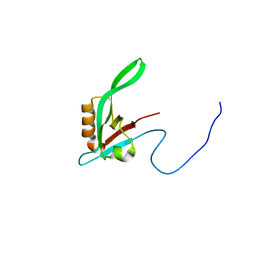 | |
1S0M
 
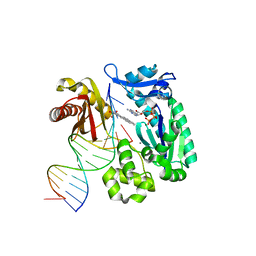 | | Crystal structure of a Benzo[a]pyrene Diol Epoxide adduct in a ternary complex with a DNA polymerase | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*T)-3', ... | | Authors: | Ling, H, Sayer, J.M, Boudsocq, F, Plosky, B.S, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-31 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1X70
 
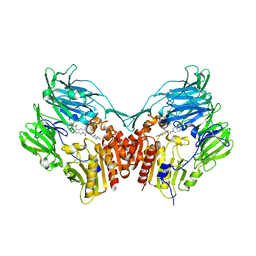 | | HUMAN DIPEPTIDYL PEPTIDASE IV IN COMPLEX WITH A BETA AMINO ACID INHIBITOR | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, D, Wang, L, Beconi, M, Eiermann, G.J, Fisher, M.H, He, H, Hickey, G.J, Leiting, B, Lyons, K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (2R)-4-Oxo-4-[3-(Trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin- 7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: A Potent, Orally Active Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
J.Med.Chem., 48, 2005
|
|
7S5J
 
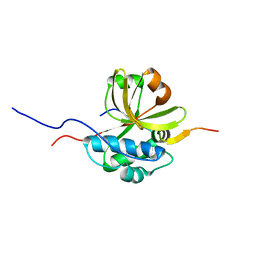 | |
7RTG
 
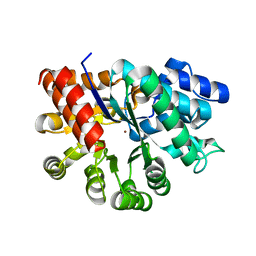 | | Crystal Structure of the Human Adenosine Deaminase 1 | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Ma, M.T, Lieberman, R.L, Blazeck, J, Jennings, M.R. | | Deposit date: | 2021-08-13 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1RXD
 
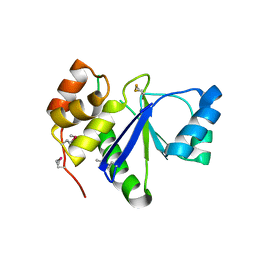 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5CPS
 
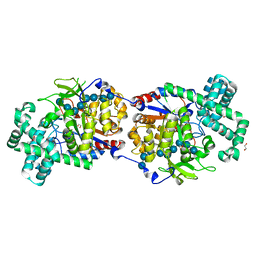 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
3LRA
 
 | | Structural Basis for Assembling a Human Tripartite Complex Dlg1-MPP7-Mals3 | | Descriptor: | Disks large homolog 1, MAGUK p55 subfamily member 7, Protein lin-7 homolog C | | Authors: | Yang, X, Xie, X, Shen, Y, Long, J. | | Deposit date: | 2010-02-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for tandem L27 domain-mediated polymerization
Faseb J., 24, 2010
|
|
5HBU
 
 | |
7S1K
 
 | | Cfr-modified Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Tsai, K, Stojkovic, V, Lee, D.J, Young, I.D, Szal, T, Vazquez-Laslop, N, Mankin, A.S, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5HD7
 
 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Mutant SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
