1MQJ
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
9GCB
 
 | | DUF4198 protein from Ideonella sakaiensis with Ni bound | | Descriptor: | MALONATE ION, NICKEL (II) ION, Nickel ECF transporter, ... | | Authors: | Franco Cairo, J.P.L, Correa, T.L.R, Offen, W.A, Nairn, A, Walton, J, Davies, G.J, Walton, P.H, Sweeney, S.T. | | Deposit date: | 2024-08-01 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Signal-strapping as a protein-sequence search method for the discovery of metalloproteins.
Nat Commun, 16, 2025
|
|
9GMU
 
 | |
1LZQ
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with an ethylenamine peptidomimetic inhibitor BOC-PHE-PSI[CH2CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Hasek, J, Dohnalek, J, Petrokova, H, Buchtelova, E, Soucek, M, Majer, P, Uhlikova, T, Konvalinka, J. | | Deposit date: | 2002-06-11 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Ethylenamine Inhibitor Binds Tightly to Both Wild Type and Mutant HIV-1 Proteases. Structure and Energy Study
J.Med.Chem., 46, 2003
|
|
9GIA
 
 | |
9GN5
 
 | | EpxF in complex with FAD from Goodfellowiella coeruleoviolacea | | Descriptor: | EpxF, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Walter, A, Kuttenlochner, W, Eisenreich, W, Groll, M, Storch, G. | | Deposit date: | 2024-08-30 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies of alpha ', beta '-Epoxyketone Synthesis by Small-Molecule Flavins and Flavoenzymes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9GHO
 
 | | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of S284A | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Creon, A, Scheer, T.E.S, Lane, T.J, Rahmani Mashhour, A, Guenther, S, Reinke, P.Y.A, Meents, A, Chapman, H.N. | | Deposit date: | 2024-08-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of S284A
To Be Published
|
|
1M51
 
 | | PEPCK complex with a GTP-competitive inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Foley, L.H, Wang, P, Dunten, P, Wertheimer, S.J. | | Deposit date: | 2002-07-06 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structures of two xanthine inhibitors bound to PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
9GHN
 
 | | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Creon, A, Scheer, T.E.S, Lane, T.J, Rahmani Mashhour, A, Guenther, S, Reinke, P.Y.A, Meents, A, Chapman, H.N. | | Deposit date: | 2024-08-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A
To Be Published
|
|
1M5N
 
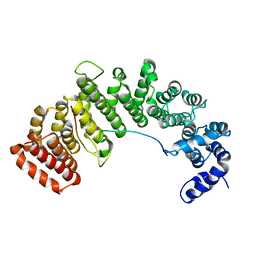 | | Crystal structure of HEAT repeats (1-11) of importin b bound to the non-classical NLS(67-94) of PTHrP | | Descriptor: | Importin beta-1 subunit, Parathyroid hormone-related protein | | Authors: | Cingolani, G, Bednenko, J, Gillespie, M.T, Gerace, L. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the recognition
of a nonclassical nuclear localization
signal by importin beta
Mol.Cell, 10, 2002
|
|
9GJV
 
 | |
7S0R
 
 | |
9GLC
 
 | |
1M5R
 
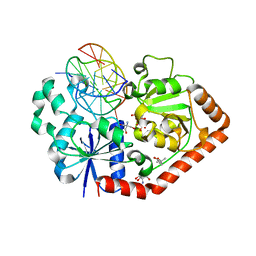 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Base-flipping mechanism for the T4 phage beta-glucosyltransferase and
identification of a transition state analog
J.Mol.Biol., 324, 2002
|
|
5FMB
 
 | |
1M6A
 
 | |
9GJX
 
 | | Bacillus licheniformis nitroreductase | | Descriptor: | 1,2-ETHANEDIOL, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Crennell, S.J, Danson, M.J, Emptage, C. | | Deposit date: | 2024-08-23 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Bacillus licheniformis nitroreductase and analysis of prodrug specificity
To Be Published
|
|
1M00
 
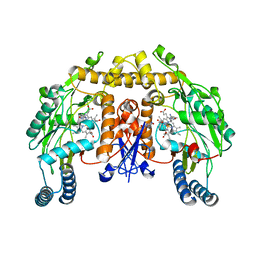 | | Rat neuronal NOS heme domain with N-butyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-BUTYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
9GN0
 
 | |
9GMJ
 
 | | The crystal structure of ManDH5 Selenomethionine derivative at 1.6 Angstroms resolution - a beta-D-Mannanase of GH5 family from Dictyoglomus thermophilium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DUF5060 domain-containing protein, GLYCEROL | | Authors: | Sivron, Y, Romano, A, Shoham, Y, Shoham, G. | | Deposit date: | 2024-08-28 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of ManDH5 Selenomethionine derivative at 1.6 Angstroms Resolution - a beta-D-Mannanase of GH5 family from Dictyoglomus thermophilium
To Be Published
|
|
1M0S
 
 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID IR21) | | Descriptor: | CITRIC ACID, Ribose-5-Phosphate Isomerase A | | Authors: | Das, K, Xiao, R, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-14 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-RIBOSE-5-PHOSPHATE ISOMERASE, IR21
TO BE PUBLISHED
|
|
1M1A
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
7S3G
 
 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
