5FX5
 
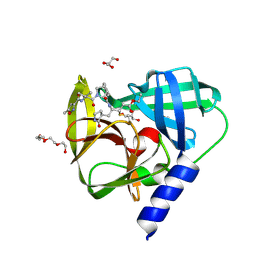 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5FX6
 
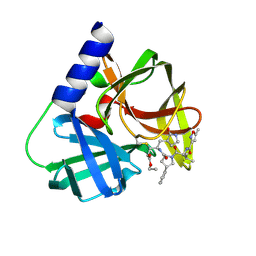 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | RHINOVIRUS 3C PROTEASE, ethyl (4R)-4-[[(2S,4S)-1-[(2S)-3-methyl-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]butanoyl]-4-phenyl-pyrrolidin-2-yl]carbonylamino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4FFZ
 
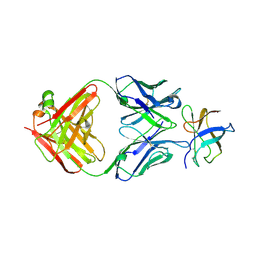 | | Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). | | Descriptor: | DENV1-E111 fab fragment (heavy chain), DENV1-E111 fab fragment (light chain), Envelope protein E | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
5FP6
 
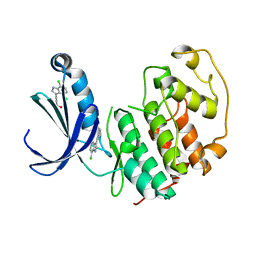 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1S97
 
 | | DPO4 with GT mismatch | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*G)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
6WBV
 
 | | Structure of human ferroportin bound to hepcidin and cobalt in lipid nanodisc | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, COBALT (II) ION, Fab45D8 Heavy Chain, ... | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
4N4S
 
 | | A Double Mutant Rat Erk2 in Complex With a Pyrazolo[3,4-d]pyrimidine Inhibitor | | Descriptor: | 3-[2-(benzyloxy)-8-methylquinolin-6-yl]-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Mitogen-activated protein kinase 1 | | Authors: | Hari, S.B, Maly, D.J, Merritt, E.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-Selective ATP-Competitive Inhibitors Control Regulatory Interactions and Noncatalytic Functions of Mitogen-Activated Protein Kinases.
Chem.Biol., 21, 2014
|
|
3MU6
 
 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
6W4S
 
 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6WK2
 
 | | SETD3 mutant (N255V) in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 2, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
1S9F
 
 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
6WK1
 
 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
5KKL
 
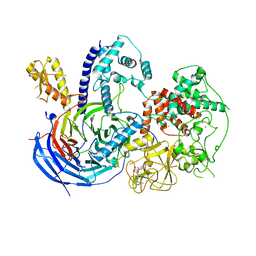 | | Structure of ctPRC2 in complex with H3K27me3 and H3K27M | | Descriptor: | ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA, Putative uncharacterized protein,Histone H3.1 peptide,Zinc finger domain-containing protein, S-ADENOSYLMETHIONINE, ... | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2016-06-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Response to Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
4EN5
 
 | | Crystal structure of fluoride riboswitch, Tl-Acetate soaked | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENA
 
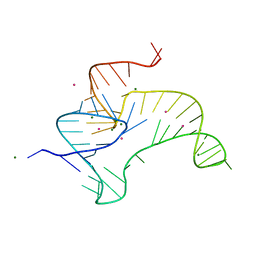 | | Crystal structure of fluoride riboswitch, soaked in Cs+ | | Descriptor: | CESIUM ION, FLUORIDE ION, Fluoride riboswitch, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENB
 
 | | Crystal structure of fluoride riboswitch, bound to Iridium | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENC
 
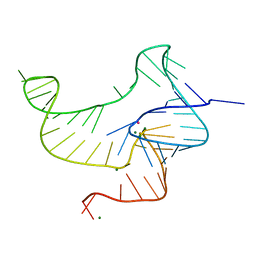 | | Crystal structure of fluoride riboswitch | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
3F2C
 
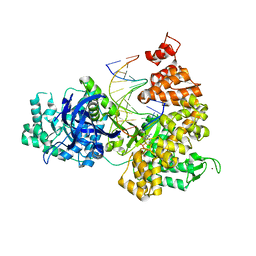 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP and Mn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6IRL
 
 | |
1S10
 
 | | Snapshots of replication through an abasic lesion: structural basis for base substitution and frameshift | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Snapshots of Replication through an Abasic Lesion; Structural Basis for Base Substitutions and Frameshifts.
Mol.Cell, 13, 2004
|
|
1S0N
 
 | | Snapshots of replication through an abasic lesion: structural basis for base substitution and frameshift | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*AP*CP*G)-3', 5'-D(*TP*AP*CP*GP*AP*CP*GP*TP*GP*AP*TP*CP*AP*GP*TP*GP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-31 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Snapshots of Replication through an Abasic Lesion; Structural Basis for Base Substitutions and Frameshifts.
Mol.Cell, 13, 2004
|
|
1S0O
 
 | | Snapshots of replication through an abasic lesion: structural basis for base substitution and frameshift | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*C)-3', 5'-D(*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', CALCIUM ION, ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-31 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Snapshots of Replication through an Abasic Lesion; Structural Basis for Base Substitutions and Frameshifts.
Mol.Cell, 13, 2004
|
|
1PIV
 
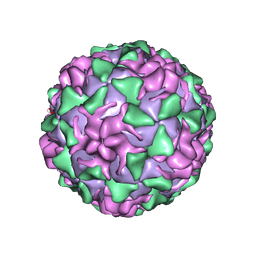 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1NG0
 
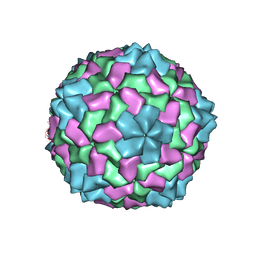 | |
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
