2C3N
 
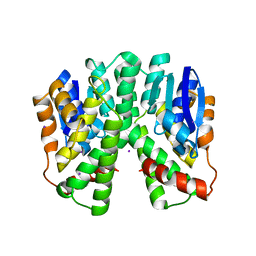 | | Human glutathione-S-transferase T1-1, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
1RTJ
 
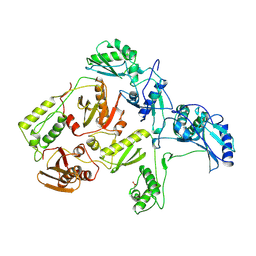 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1D4I
 
 | | HIV-1 protease in complex with the inhibitor BEA425 | | Descriptor: | 2,5-DIBENZYLOXY-3-HYDROXY-HEXANEDIOIC ACID BIS-[(2-HYDROXY-INDAN-1-YL)-AMIDE], HIV-1 PROTEASE | | Authors: | Unge, T. | | Deposit date: | 1999-10-04 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors.
Eur.J.Biochem., 270, 2003
|
|
3TGQ
 
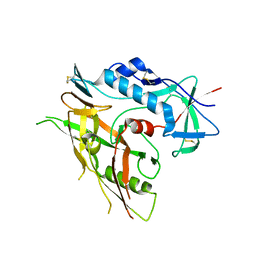 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2GMW
 
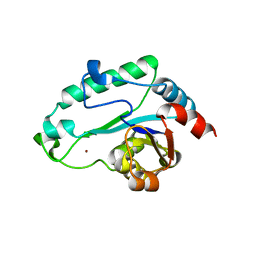 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli. | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Zhang, K, DeLeon, G, Wright, G.D, Junop, M.S. | | Deposit date: | 2006-04-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
2C8N
 
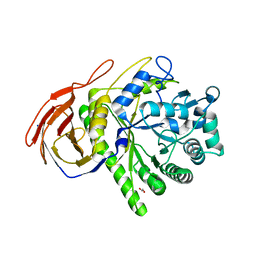 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,3-linked arabinoside of xylobiose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-3)-alpha-D-xylopyranose | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
3TAM
 
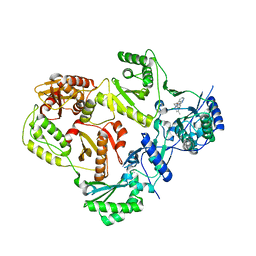 | |
3P71
 
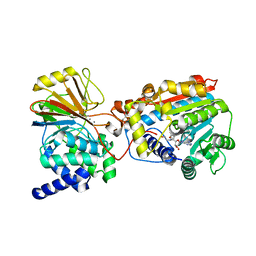 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
3OQD
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OX6
 
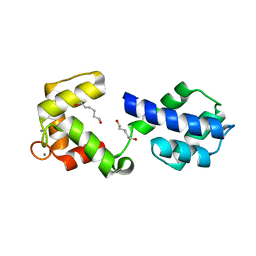 | |
1TDL
 
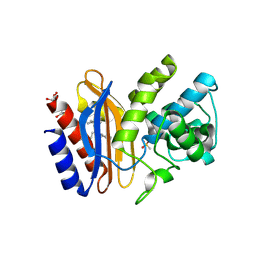 | | Structure of Ser130Gly SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sun, T, Bethel, C.R, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-05-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant
Biochemistry, 43, 2004
|
|
3GO4
 
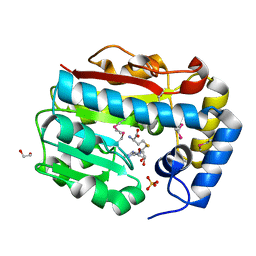 | |
2M9R
 
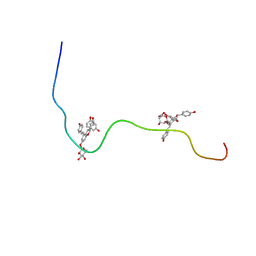 | |
2L94
 
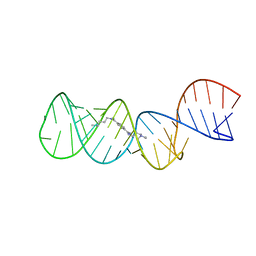 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
1D4J
 
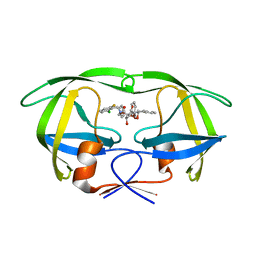 | | HIV-1 protease in complex with the inhibitor MSL370 | | Descriptor: | 2,5-DIBENZYLOXY-3,4-DIHYDROXY-HEXANEDIOIC ACID 2-CHLORO-6-FLUORO-BENZYLAMIDE (2-HYDROXY-INDAN-1- YL)-AMIDE, HIV-1 PROTEASE | | Authors: | Unge, T. | | Deposit date: | 1999-10-04 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors.
Eur.J.Biochem., 270, 2003
|
|
2M9S
 
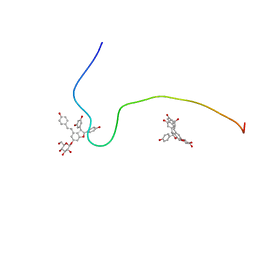 | |
1LRH
 
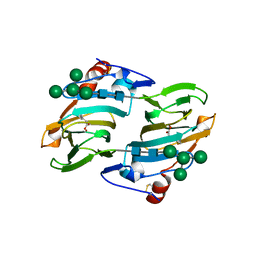 | | Crystal structure of auxin-binding protein 1 in complex with 1-naphthalene acetic acid | | Descriptor: | NAPHTHALEN-1-YL-ACETIC ACID, ZINC ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Woo, E.J, Marshall, J, Bauly, J, Chen, J.-G, Venis, M, Napier, R.M, Pickersgill, R.W. | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of auxin-binding protein 1 in complex with auxin.
EMBO J., 21, 2002
|
|
3U82
 
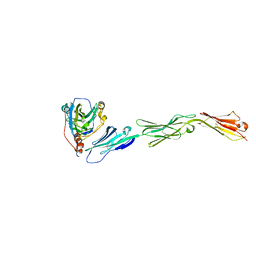 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
2VJ9
 
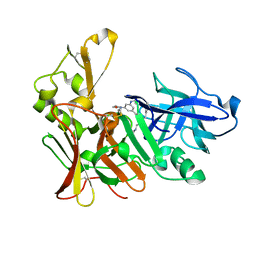 | | Human BACE-1 in complex with N-((1S,2R)-3-(cyclohexylamino)-2-hydroxy- 1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl) benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-(cyclohexylamino)-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VJ7
 
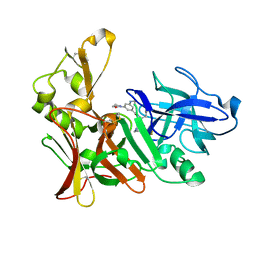 | | Human BACE-1 in complex with 3-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VIZ
 
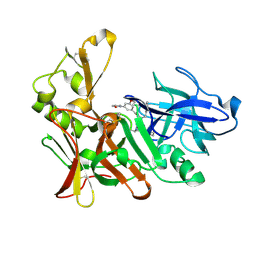 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(2-oxo- 1-pyrrolidinyl)-5-(propyloxy)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)-5-propoxybenzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 1: Identification of Novel Hydroxy Ethylamines (Heas).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5T3X
 
 | |
3EL1
 
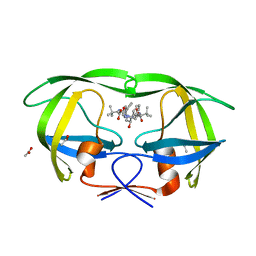 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EKY
 
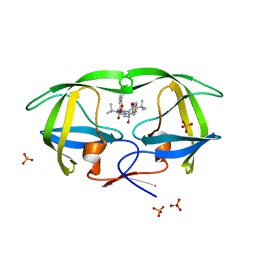 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
