6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
6NEL
 
 | | 4-(2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzoic acid bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 4-[2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl]benzoic acid, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl and Arylalkyl Substituted 3-Hydroxypyridin-2(1H)-ones: Synthesis and Evaluation as Inhibitors of Influenza A Endonuclease.
Chemmedchem, 14, 2019
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
8OR4
 
 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
5O8K
 
 | |
7LUV
 
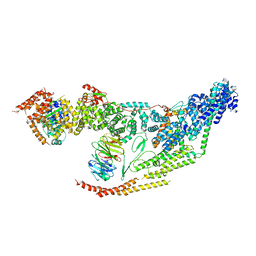 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
3BRD
 
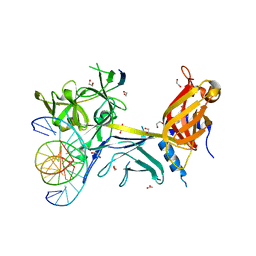 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
3GPG
 
 | |
5TXL
 
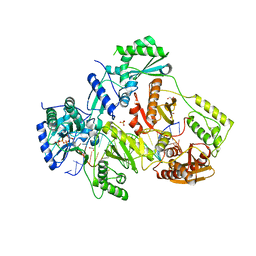 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG) P*CP*GP*CP*CP*GP)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXN
 
 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXM
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
6SN1
 
 | | Crystal structure of the human INTS13-INTS14 complex | | Descriptor: | Integrator complex subunit 13, Integrator complex subunit 14, SULFATE ION | | Authors: | Jonas, S, Sabath, K, Staeubli, M.L. | | Deposit date: | 2019-08-23 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module.
Nat Commun, 11, 2020
|
|
3TG4
 
 | | Structure of SMYD2 in complex with SAM | | Descriptor: | GLYCEROL, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
2BE2
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with R221239 | | Descriptor: | 4-(3,5-DIMETHYLPHENOXY)-5-(FURAN-2-YLMETHYLSULFANYLMETHYL)-3-IODO-6-METHYLPYRIDIN-2(1H)-ONE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Himmel, D.M, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Nguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1J5O
 
 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2B5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R165481 | | Descriptor: | (2E)-3-{3-[(5-ETHYL-3-IODO-6-METHYL-2-OXO-1,2-DIHYDROPYRIDIN-4-YL)OXY]PHENYL}ACRYLONITRILE, MANGANESE (II) ION, Reverse transcriptase P51 SUBUNIT, ... | | Authors: | Himmel, D.H, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Mguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
2BAN
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R157208 | | Descriptor: | 5-ETHYL-3-[(2-METHOXYETHYL)METHYLAMINO]-6-METHYL-4-(3-METHYLBENZYL)PYRIDIN-2(1H)-ONE, MANGANESE (II) ION, Reverse transcriptase P51 subunit, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
4QBB
 
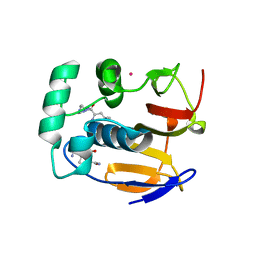 | | Structure of the foot-and-mouth disease virus leader proteinase in complex with inhibitor (N~2~-[(3S)-4-({(2R)-1-[(4-CARBAMIMIDAMIDOBUTYL)AMINO]-4-METHYL-1-OXOPENTAN-2-YL}AMINO)-3-HYDROXY-4-OXOBUTANOYL]-L-ARGINYL-L-PROLINAMIDE) | | Descriptor: | Leader protease, N~2~-[(3S)-4-({(2R)-1-[(4-carbamimidamidobutyl)amino]-4-methyl-1-oxopentan-2-yl}amino)-3-hydroxy-4-oxobutanoyl]-L-arginyl-L-prolinamide, PHOSPHATE ION, ... | | Authors: | Grishkovskaya, I, Steinberger, J, Cencic, R, Juliano, M.A, Juliano, L, Skern, T. | | Deposit date: | 2014-05-07 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Foot-and-mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage.
Virology, 468-470C, 2014
|
|
3LKW
 
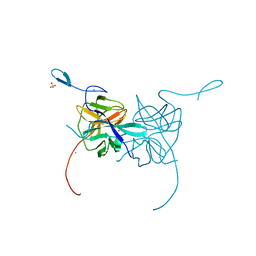 | | Crystal Structure of Dengue Virus 1 NS2B/NS3 protease active site mutant | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chandramouli, S, Joseph, J.S, Daudenarde, S, Gatchalian, J, Cornillez-Ty, C, Kuhn, P. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family.
J.Virol., 84, 2010
|
|
7OZR
 
 | |
6ERO
 
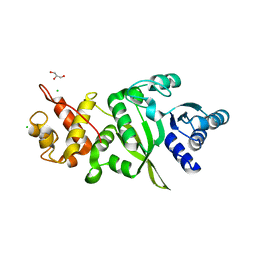 | | Structure of human TFB2M | | Descriptor: | CHLORIDE ION, Dimethyladenosine transferase 2, mitochondrial,Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
3L6P
 
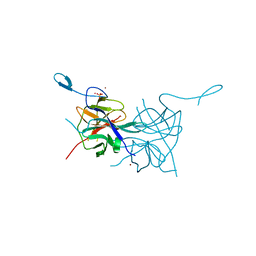 | | Crystal Structure of Dengue Virus 1 NS2B/NS3 protease | | Descriptor: | CADMIUM ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Chandramouli, S, Joseph, J.S, Daudenarde, S, Gatchalian, J, Cornillez-Ty, C, Kuhn, P. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family.
J.Virol., 84, 2010
|
|
3VKW
 
 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
8EQB
 
 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
