6GA3
 
 | | Bacteriorhodopsin, 33 ms state, ensemble refinement | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAC
 
 | | BACTERIORHODOPSIN, 490 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GA5
 
 | | Bacteriorhodopsin, 3 ps state, REAL-SPACE REFINEMED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAF
 
 | | BACTERIORHODOPSIN, 590 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
1GCT
 
 | |
1H51
 
 | | Oxidised Pentaerythritol Tetranitrate Reductase (SCN complex) | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, THIOCYANATE ION | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H50
 
 | | Structure of Pentaerythritol Tetranitrate Reductase and complexes | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
5JR5
 
 | | RHCC in Complex with Elemental Sulfur | | Descriptor: | SULFATE ION, Tetrabrachion, octathiocane | | Authors: | McDougall, M, Meier, M, Stetefeld, J. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Archaea S-layer nanotube from a "black smoker" in complex with cyclo-octasulfur (S8 ) rings.
Proteins, 85, 2017
|
|
3BNI
 
 | | Crystal structure of TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | Putative TetR-family transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Osipiuk, J, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of TetR-family transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
3L03
 
 | | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II peptide and Estetrol (Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol) | | Descriptor: | (14beta,15alpha,16alpha,17alpha)-estra-1,3,5(10)-triene-3,15,16,17-tetrol, CHLORIDE ION, Estrogen receptor, ... | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain
in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II
peptide and Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol
To be Published
|
|
6CRD
 
 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) with tetrabrachion (TB) domain stalk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tetrabrachion,Neuraminidase, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4W5I
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-phenyl-1,2,3,4,5,6-hexahydro-1,6- naphthyridin-5-one | | Descriptor: | 1-methyl-7-phenyl-2,3,4,6-tetrahydro-1,6-naphthyridin-5(1H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
4JKI
 
 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with ZD9331, Formylphosphate, and ADP | | Descriptor: | 2,7-dimethyl-6-[(prop-1-yn-1-ylamino)methyl]quinazolin-4(3H)-one, ADENOSINE-5'-DIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2013-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors.
Protein Sci., 21, 2012
|
|
4JIM
 
 | |
4JJK
 
 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with Folate | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, FOLIC ACID, ... | | Authors: | Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors.
Protein Sci., 21, 2012
|
|
4JJZ
 
 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with ADP and Formylphosphate | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ADENOSINE-5'-DIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors.
Protein Sci., 21, 2012
|
|
4WTY
 
 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
4WU3
 
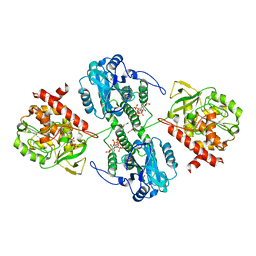 | | Structure of the PTP-like myo-inositol phosphatase from Mitsuokella multacida in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, MYO-INOSITOL PHOSPHOHYDROLASE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
4X6N
 
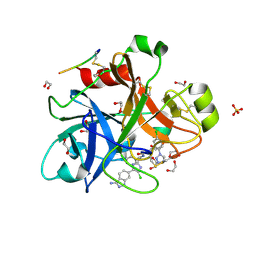 | |
4X6O
 
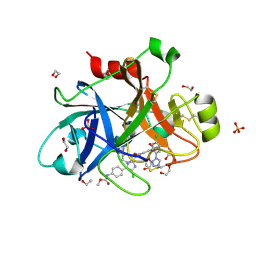 | |
4X48
 
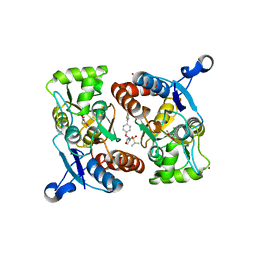 | | Crystal structure of GluR2 ligand-binding core | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2014-12-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
6V4L
 
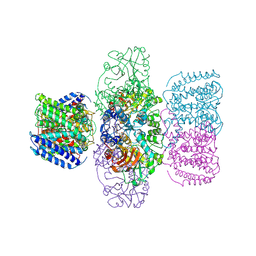 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6V4J
 
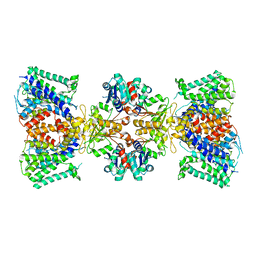 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6V4K
 
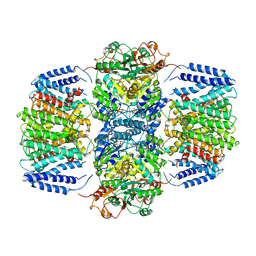 | | Structure of TrkH-TrkA in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Potassium transporter peripheral membrane component, Trk system potassium uptake protein | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.53004146 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
3FST
 
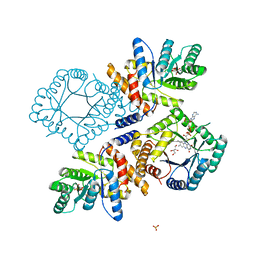 | |
